Bot. Bull. Acad. Sin. (2004) 45: 23-31
Lu et al. — Coniothyrium minitans xylanase in Arabidopsis
Segregation patterns for integration and expression of Coniothyrium minitans xylanase gene in Arabidopsis thaliana transformants
Z.-X. Lu, A. Laroche*, and H.C. Huang
Agriculture and Agri-Food Canada, Lethbridge Research Centre, P.O. Box 3000, Lethbridge, Alberta T1J 4B1, Canada
(Received October 8, 2002; Accepted August 26, 2003)
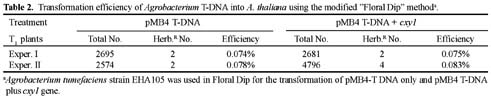
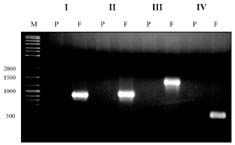
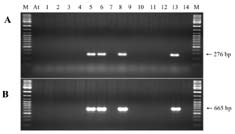
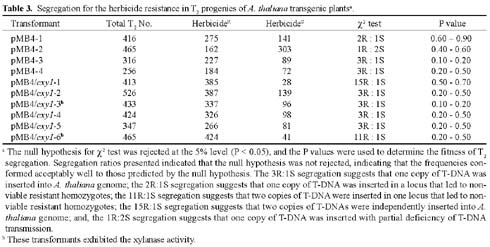
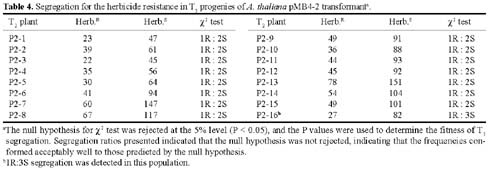
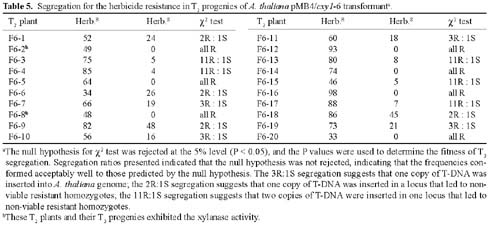
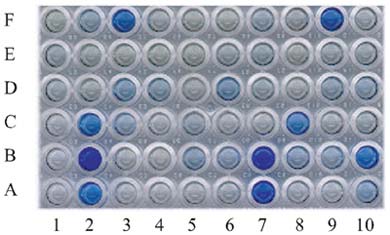
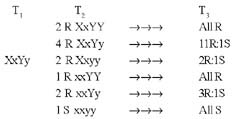
Abstract. A xylanase gene cxy1, isolated from the mycoparasitic fungus Coniothyrium minitans, has been transferred into Arabidopsis thaliana by Agrobacterium-mediated transformation. Ten A. thaliana transformants with herbicide resistance were selected, and the transformation efficiency of the modified "Floral Dip" method was about 0.08 %. Most transformants had one or two copies of T-DNA inserts with 3R:1S or 15R:1S segregation ratios while high proportions of susceptible progenies were also observed in some transgenic plants. The pMB4-2 transformant segregated as 1R:2S ratio in its T2 and T3 generations, suggesting that only one copy of T-DNA was integrated into this line's genome and the insertion likely interrupted a gene important or essential to its gametophytic development or alternatively, affected the transmission of such gene. The genetic analysis of the pMB4/cxy1-6 transformant indicated that two copies of T-DNAs were inserted independently into this line's genome, in which the resistant homozygotes on one locus were non-viable. The RBB-Xylan assay for six pMB4/cxy1 transformants indicated that two lines (pMB4/cxy1-3 and pMB4/cxy1-6) expressed the cxy1 gene, and variations for the xylanase activity were observed among T2 progenies of pMB4/cxy1-6 transformant.
Keywords: Agrobacterium-mediated transformation; Arabidopsis thaliana; Coniothyrium minitans; Xylanase gene.
Introduction
Xylan is a complex polysaccharide which consists of a backbone of xylose residues linked by b-1,4-glycosidic bonds. Xylan fibers constitute a significant portion of plant hemi-cellulose and contribute to the strength of plant secondary walls (Thomson, 1993). The hydrolysis of xylan by xylanases (E.C. 3.2.1.8) produced by fibrolytic bacteria and fungi is necessary in the degradation of plant tissue. Xylanolytic enzymes have numerous applications in industry and biotechnology, such as in woody biopulping processes (Bieley, 1985), forage fibre digestions (Gilbert and Hazelwood, 1991), and agricultural waste degradation.
Agrobacterium-mediated gene transfer is the most commonly used technique for plant transformation (Zambryski, 1992; Zupan and Zambryski, 1995). The `Agrobacterium Vacuum Infiltration', a non-tissue culture approach for in planta transformation, was first achieved in Arabidopsis thaliana (L.) Heynh. (Feldmann and Marks, 1987; Bechtold et al., 1993; Chang et al., 1994; Katavic et al., 1994). Later, a relatively simplified protocol called the "Floral Dip" was described by Clough and Bent (1998). However, it still re
quires sterile conditions for seed germination and transformant selection. Recently, we successfully modified this method by germinating A. thaliana seeds and selecting transgenic seedlings in Cornell Peat-Lite Mix in plastic pots under greenhouse conditions.
Coniothyrium minitans Campbell is a mycoparasite which attacks the sclerotia (Huang, 1977; Huang and Kokko, 1987) and hyphae (Huang and Hoes, 1976; Huang and Kokko, 1988) of Sclerotinia sclerotiorum (Lib.) de Bary, an important fungal pathogen of higher plants (Purdy, 1979). A novel xylanase gene cxy1 has been isolated from C. minitans (Laroche et al., 2000) and expressed in Pichia pastoris (Guilliermond) Phaff (Lu et al., 1999). The objective of our study is to explore the impacts of cxy1 gene for resistance to S. sclerotiorum in plants and for improvement of forage utilization in animal production. This report describes the experimental results for cxy1 integration and expression in A. thaliana.
Materials and Methods
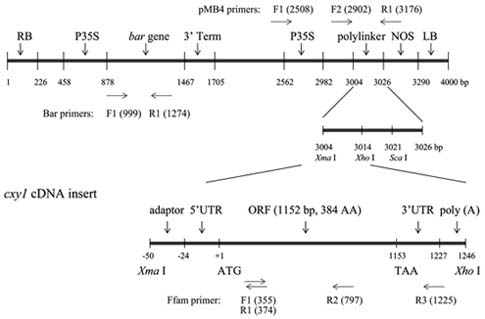
Construction of Ti Plasmid
The Agrobacterium tumefaciens (Smith & Townsend) Conn strain EHA105 was provided by Dr. F. Eudes (Lethbridge Research Centre, Agriculture and Agri-Food Canada), and the binary vector pMB4 was obtained from "The Baker Lab" at the Gene Expression Center (Berkeley, CA). Strain EHA105 already contains the helper vector,
Lethbridge Research Centre, Canada, Contribution No. 387-01021.
*Corresponding author. Tel: + 1 (403) 317-2224; Fax: + 1 (403) 382-3156; E-mail: laroche@agr.gc.ca