Bot. Bull. Acad. Sin. (2004) 45: 61-68
Lee et al. — Comparison of Aspergillus flavus and A. oryzae by amplified fragment length polymorphism
Comparison of Aspergillus flavus and Aspergillus oryzae by amplified fragment length polymorphism
Chao-Zong Lee, Guey-Yuh Liou, and Gwo-Fang Yuan*
Bioresource Collection and Research Center, Food Industry Research and Development Institute, P.O. Box 246, Hsinchu, Taiwan 300, Republic of China
(Received April 25, 2003; Accepted October 31, 2003)
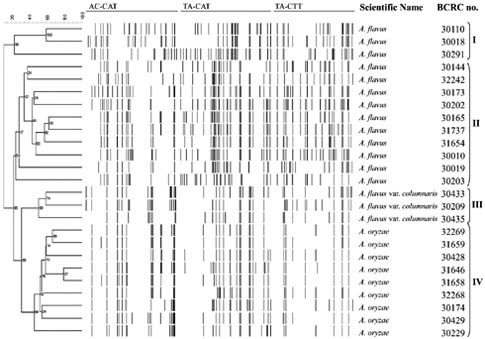
Abstract. The aflatoxin producer Aspergillus flavus and the koji mold Aspergillus oryzae are morphologically similar species that belong to the Aspergillus section Flavi. We used amplified fragment length polymorphism (AFLP) to differentiate these two species. In this study, we tested thirteen A. flavus, nine A. oryzae, and three A. flavus var. columnaris strains. DNA fragment profiles amplified with each of three selective primer pairs displayed similar patterns for the various A. flavus strains. Different patterns were observed with these primer pairs for the A. oryzae strains. We combined these data to increase the grouping of the various strains within each species and to distinguish between A. flavus and A. oryzae. By unweighted pair group method with arithmetic average (UPGMA) analysis, the AFLP data obtained from the three selective primer pairs, EcoRI-AC/MseI-CAT, EcoRI-TA/MseI-CAT, and EcoRI-TA/MseI-CTT, differentiated A. flavus from A. oryzae successfully. Three strains of A. flavus received from ATCC grouped outside of the other A. flavus strains we examined. The morphology of these isolates and our results indicated those strains were originally misidentified and they should be classified as A. parasiticus. We found that the AFLP technique is a reliable and reproducible tool with the ability to differentiate A. flavus from A. oryzae and should be generally useful in distinguishing between closely related species or strains.
Keywords: AFLP; Aspergillus flavus; Aspergillus oryzae.
Introduction
The aflatoxigenic species Aspergillus flavus Link and A. parasiticus Speare can produce the potent carcinogen aflatoxin and can pose a significant human health threat. The nonaflatoxigenic species A. oryzae (Ahlburg) Cohn and A. sojae Sakaguchi & Yamada are widely used for the production of food-grade amylase and the fermentation of sake, miso and soy sauce. All four species belong to the Aspergillus section Flavi and have many phenotypic similarities. Current methods for the identification of these species still depend primarily on cultural and morphological characteristics. However, it is often difficult to differentiate these species because the phenotypic differences are not distinct and are easily affected by the environment and are also confused by the high degree of intra- and interspecies variation.
Because of the economic value of members of the Aspergillus section Flavi and the importance of differentiating them, several molecular methods have been developed to distinguish these strains. DNA relatedness of these species in Aspergillus section Flavi was studied by Kurtzman et al. (1986) with the DNA reassociation method. The Cot value calculation results showed 100% related
ness between A. flavus and A. oryzae. Similarly, A. parasiticus and A. sojae were 91% related. The homology between these two groups was 70%. Therefore Kurtzman et al. suggested the four taxa be regarded as varieties of A. flavus. A medium containing bleomycin has been used to distinguish A. parasiticus from A. sojae (Klich and Mullaney, 1989; Yuan et al., 1995). The random amplified polymorphic DNA (RAPD) method has been used to differentiate A. parasiticus from A. sojae with three screened decamer primers (Yuan et al., 1995), but the same primers could not separate A. flavus from A. oryzae (Yuan et al., unpubl.). The sequence of a regulatory gene for the aflatoxin biosynthetic pathway was analyzed, and distinct DNA sequences within the 5' untranslated region and the zinc- finger domain were found in limited strains of A. flavus, A. oryzae, A. parasiticus, and A. sojae (Chang et al., 1995). PCR-amplified regions of ribosomal internal transcribed spacers have also been used to differentiate species of Aspergillus section Flavi with the single-strand conformation polymorphism (SSCP) method (Kumeda and Asao, 1996). The SSCP results indicated that 67 of the 68 test strains could be classified into four groups as A. flavus/A. oryzae, A. parasiticus/A. sojae, A. tamari, and A. nomius. Sixteen strains of A. flavus or A. oryzae could not be clearly separated with the sequence of ITS1-5.8S-ITS2 region of ribosomal DNA (Lin et al., 1995). Restriction site polymorphism and DNA sequences of protein-encoding genes were analyzed, and 31 A. flavus strains could be separated into two distinct groups. Five
*Corresponding author. Gwo-Fang Yuan, P.O. Box 246, Hsinchu, Taiwan 300, Republic of China. Tel: 886-3-5223191-580; Fax: 886-3-5214016; E-mail: gfy@firdi.org.tw