380
Botanical Studies, Vol. 50, 2009
The expressions of most plant EGases are tightly regulated, suggesting that they play some critical roles in plant development. Some EGases, i.e. avocado CEL1, pepper CX1 and tomato Cel2, are expressed specifically during fruit ripening (Christoffersen et al. , 1 984; Lashbrook et al., 1994; Ferrarese et al., 1995). The EGases, i.e. bean BAC1, soybean SAC1 and elder JET1, are found to be associated with abscission (Tucker et al., 1988; Kemmerer and Tucker, 1994; Taylor et al., 1994). Some EGases are proposed to be involved in cell elongation and growth rate, like Arabidopsis Cel1, which is highly expressed in the elongating stem and root, and tomato Cel4, which is highly expressed in the growing etiolated hypocotyls, pistils, and expanding leaves (Milligan and Gasser, 1995; Brummell et al., 1997a; Sham et al., 1997).
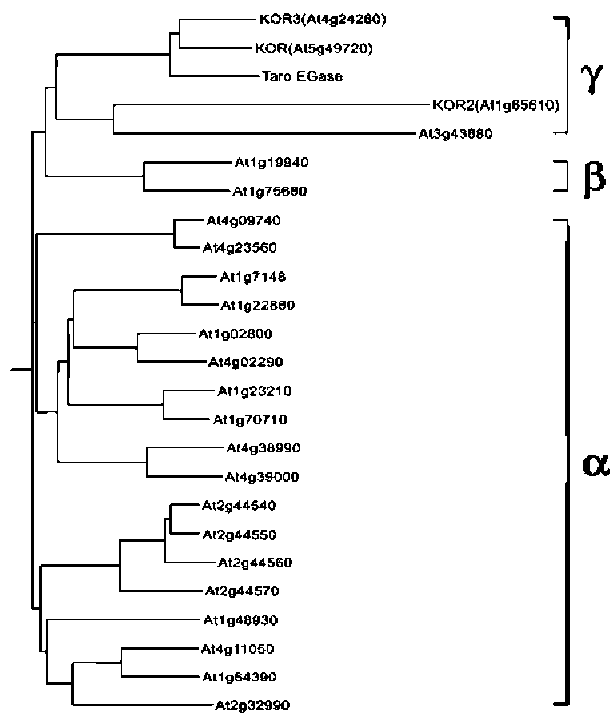
Another group of membrane-anchor EGases, located at the plasma membrane, is also required for normal cellulose formation (Brummell et al., 1997a; Nicol et al., 1998; Del Campillo, 1999; Molhoj et al., 2001a). This group includes the Arabidopsis KOR gene, which was identified by the dwarf mutant KORRIGAN and found to be involved in cytokinesis and cell elongation (Nicol et al., 1998; Lane et al., 2001). Moreover, KOR has been postulated to trim sterol residues from nascent glucan or out-of-register glucan as a primer for the elongation of microfibril (Peng et al., 2002). However, the KOR protein could not be associated with cellulose synthase complexes during microfibril elongation (Zuo et al., 2000; Szyjanowicz et al., 2004), and kor mutants contain normal amounts of sterol glycoside (Robert et al., 2004). This suggested that the action of KOR might be indirect during microfibril elongation, and the role of KOR requires more experimental evidence to be defined.
Although plant EGases were reported to be related to ripening, abscission, and cell wall metabolism, the detailed molecular mechanisms of these enzymes are still unclear. Transformation of Arabidopsis Cel1 conferred on plants an altered structure or morphology, i.e. improved growth rate, a greater biomass, or modified fibers (Shoseyov et al., 2001). Cloning of EGases will not only be critical then for studying these subjects, it is also likely to be useful in the molecular breeding of crops. In our previous work, a partial EGase cDNA of 1.8 kb was obtained unpurposely from taro (Colocasia esculenta var. esculenta) during the gene cloning process for soluble starch synthase. To further understand its functions, the full-length cDNA was isolated, characterized, and expressed, and the temporal expression profiles in the tuber and leaf tissues were also described.
MATERIALS AND METHODS Plant material
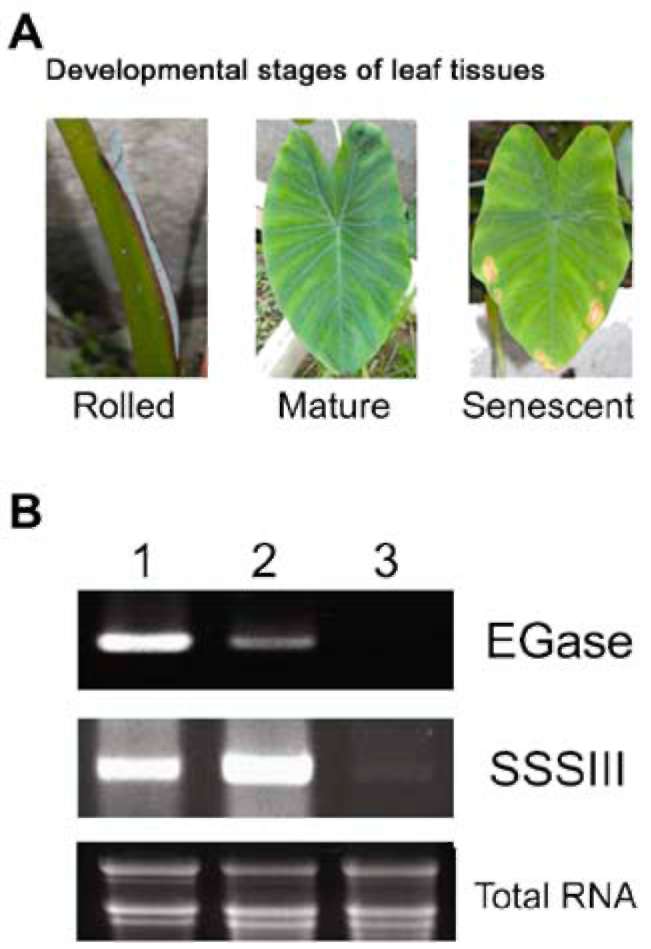
Taro (Colocasia esculenta var. esculenta) tubers were harvested during developmental stages based on their
fresh weight (i.e. 100 g, 300 g, 600 g, and 1000 g) and leaf
growth (i.e., young [rolled], mature, and senescent). The
collected leaves and tubers were frozen immediately in liquid nitrogen, lyophilized, and then stored at -20°C until required.
RNA isolation
RNA was prepared by the method described previously (Lin and Jeang, 2005). The quality and quantity of RNA were determined by spectrophotometry and agarose gel electrophoresis (Sambrook and Russell, 2001). Finally, RNA was stored at -70°C until required. The mRNA for gene cloning was purified with OligotexTM (Qiagen, Valencia, CA) according to the manufacturer's protocol.
Taro EGase cDNA cloning
A s c hema tic re pres entatio n o f our clo ni ng strategy is shown in Figure 1A. The full-length cDNA of taro EGase was cloned by combining RT-PCR and 5'-RACE. 3'-RACE using the primers
P 1 ( G C C G T T G G A C T T AT C G T G C ) a n d P 2 (GAC TC GAGT C GAC A T C G) using the method
described by Frohman et al. (1 988). The template used in 5'-RACE was prepared with oligo-dT primer by a SMART™ RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA) following the
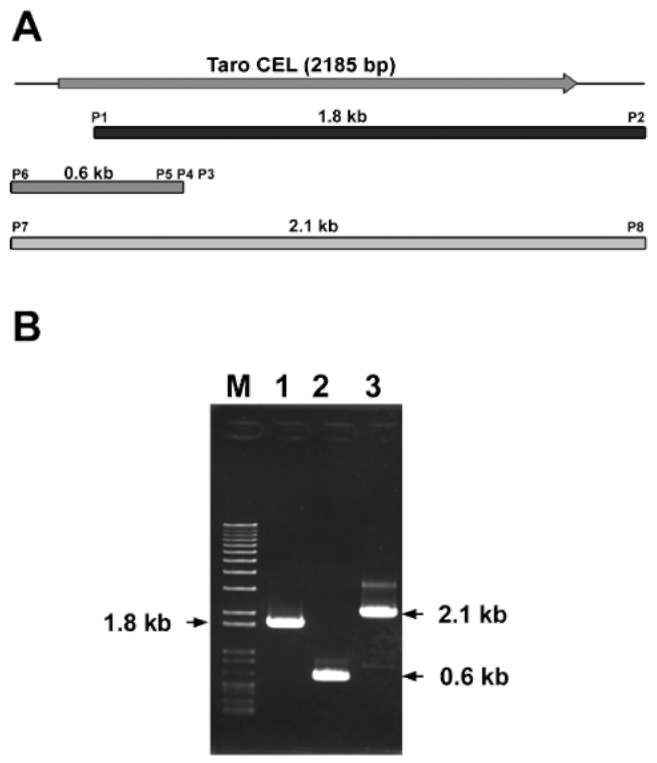
Figure 1. (A) Strategy for taro EGase cDNA cloning. The topmost diagram shows the full length cDNA. The boxes represent cDNAs amplified by RT-PCR and RACE; (B) Agarose gel electrophoresis of RACE and RT-PCR products of EGase. The partial EGase cDNAs were amplified by RT-PCR using P1 and P2 primers (lane 1); The product of 5' SMART- RACE was amplified using P3, P4, P5 and P6 primers (lane 2); The full-length of EGase cDNA was generated by RT-PCR using P7 and P8 primers (lane 3); Lane M: 100 bp ladder (MBI).