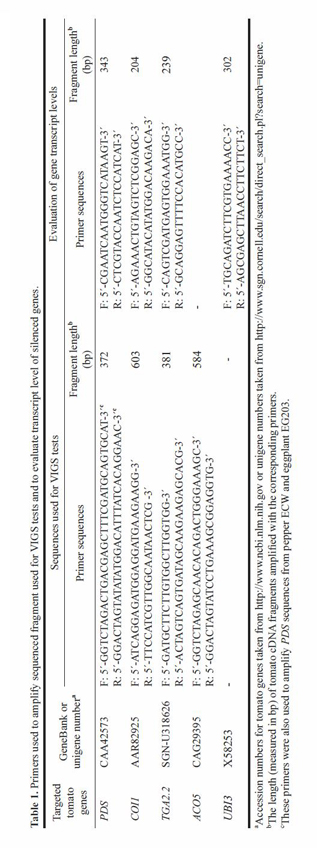
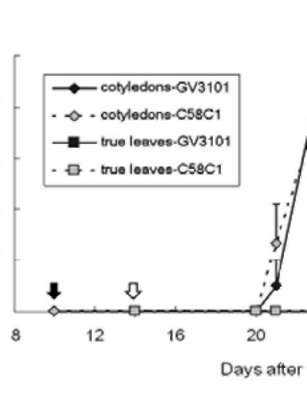
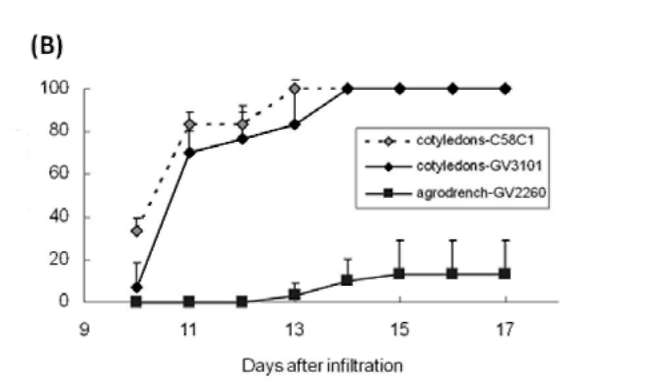
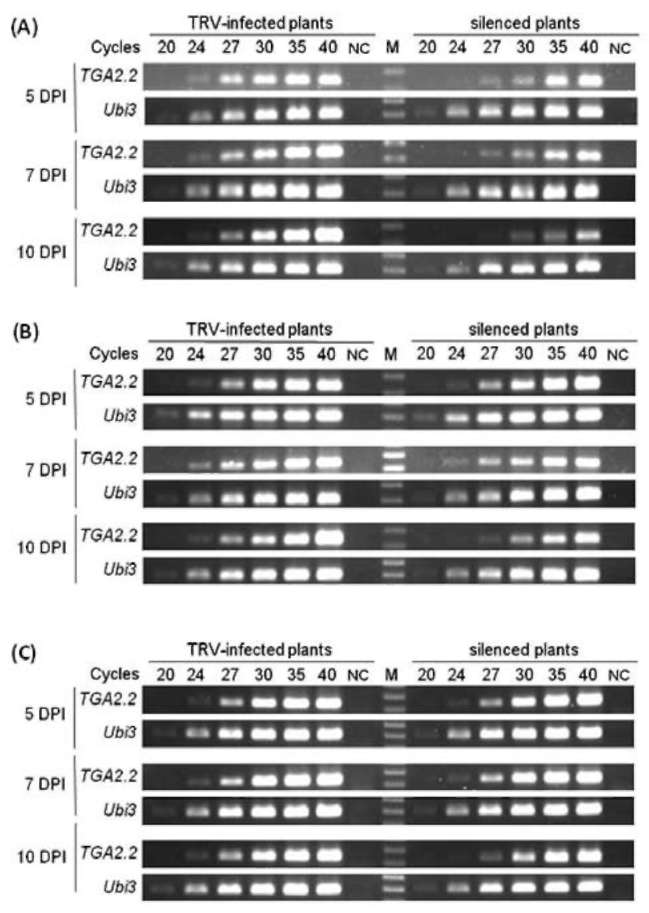
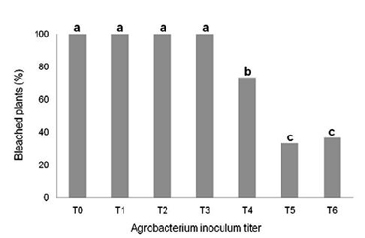
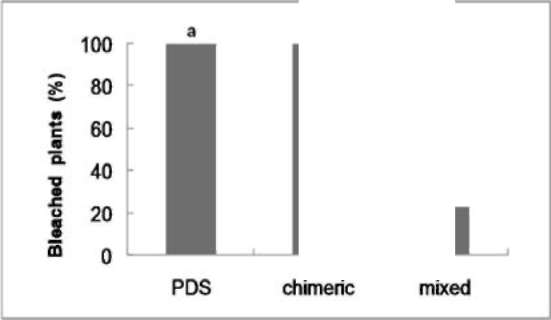
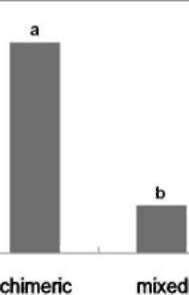
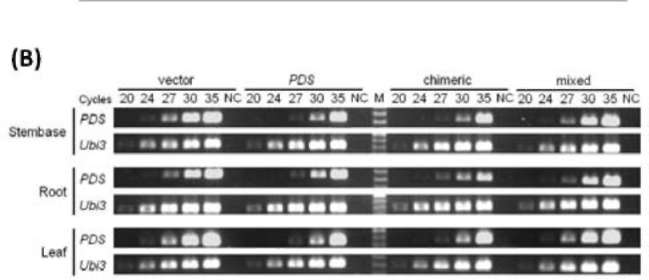
|
|
and C.P. Cheng. 2009. Tomato resistance to bacterial wilt involves ethylene, salicylic acid and mitogen-associated protein kinase related defense pathways. Physiol. Plant. (In
press)
Denny, T.P. 2006. Plant pathogenic Ralstonia species. In S.S. Gnanamanickam (ed.), Plant-Associated Bacteria. Springer, Dordrecht, The Netherlands, pp. 573-644.
Deslandes, L., J. Olivier, F. Theulieres, J. Hirsch, D.X. Feng, P. Bittner-Eddy, J. Beynon, and Y. Marco. 2002. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci.
USA 99: 2404-2409.
Ekengren, S.K., Y. Liu, M. Schiff, S.P. Dinesh-Kumar, and G.B. Martin. 2003. Two MAPK cascades, NPR1, and TGA
transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36: 905-917.
Ghazala, W. and M. Varrelmann. 2007. Tobacco rattle virus 29K movement protein is the elicitor of extreme and hypersensitive-like resistance in two cultivars of Solanum tuberosum. Mol. Plant-Microbe Interact. 20: 1396-1405.
Giritch, A., S. Marillonnet, C. Engler, G. van Eldik, J.
Botterman, V. Klimyuk, and Y. Gleba. 2006. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl.
Acad. Sci. USA 103: 14701-14706.
Grimault, V., G. Anais, and P. Prior. 1994. Distribution of Pseudomonas solanacearum in the stem tissues of tomato plants with different levels of resistance to bacterial wilt.
Plant Pathol. 43: 663-668. Hartl, M., H. Merker, D.D. Schmidt, and I.T. Baldwin. 2008.
Optimized virus-induced gene silencing in Solanum nigrum reveals the defensive function of leucine aminopeptidase against herbivores and the shortcomings of empty vector controls. New Phytol. 179: 356-365.
Hernandez-Blanco, C., D.X. Feng, J. Hu, A. Sanchez-Vallet, L. Deslandes, F. Llorente, M. Berrocal-Lobo, H. Keller, X. Barlet, C. Sanchez-Rodriguez, L. K. Anderson, S. Somerville, Y. Marco, and A. Molina. 2007. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19:
890-903.
Hirsch, J. , L. Deslandes, D.X. Feng, C. Balague, and Y. Marco. 2002. Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum.
Phytopathology 92: 1142-1148. Hwang, K.Y., I.C. Chou, Y.M. Lin, and C.P. Cheng. 2004. Plant
age-related resistance to Ralstonia solanacearum, the causal
agent of bacterial wilt. J. Gene Mol. Biol. 15: 108-115. Kunkel, B.N. and D.M. Brooks. 2002. Cross talk between
signaling pathways in pathogen defense. Curr. Opin. Plant
Biol. 5: 325-331.
Kandoth, P.K., S. Ranf, S.S. Pancholi, S. Jayanty, M.D. Walla, W. Miller, G.A. Howe, D.E. Lincoln, and J.W. Stratmann.
|
2007. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3
function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 104:
12205-12210.
Li, L., Y. Zhao, B.C. McCaig, B.A. Wingerd, J. Wang, M.E.
Whalon, E. Pichersky, and G.A. Howe. 2004. The tomato
homolog of CORONATINE-INSENSITIVE1 is required for
the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development.
Plant Cell 16: 126-143.
Lin, W.C., C.F. Lu, J.W. Wu, M.L. Cheng, Y.M. Lin, N.S. Yang, L. Black, S.K. Green, J.F. Wang, and C.P. Cheng. 2004.
Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13: 567-581.
Lin, Y.M., I.C. Chou, J.F. Wang, F.I. Ho, Y.J. Chu, P.C. Huang, D.K. Lu, H.L. Shen, M. Elbaz, S.M. Huang, and C.P.
Cheng. 2008. Transposon mutagenesis reveals differential pathogenesis of Ralstonia solanacearum on tomato and
Arabidopsis. Mol. Plant-Microbe Interact. 21: 1261-1270. Liu, Y., M. Schiff, and S.P. Dinesh-Kumar. 2002. Virus-induced
gene silencing in tomato. Plant J. 31: 777-786.
Ossowski, S., R. Schwab, and D. Weigel. 2008. Gene silencing in plants using artificial microRNAs and other small RNAs.
Plant J. 53: 674-690.
Pieterse, C.M. and M. Dicke. 2007. Plant interactions with microbes and insects: from molecular mechanisms to
ecology. Trends Plant Sci. 12: 564-569.
Prior, P., V. Grimault, and J. Schmit. 1994. Resistance to bacterial wilt (Pseudomonas solanacearum) in tomato: present status and prospects. In A.C. Hayward and G.L. Hartman (eds.), Bacterial Wilt: The disease and its causative agent, Pseudomonas solanacearum. CAB International,
Wallingford, pp. 209-223.
Robatzek, S., P. Bittel, D. Chinchilla, P. Kochner, G. Felix, S.H. Shiu, and T. Boller. 2007. Molecular identification
and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant
Mol. Biol. 64: 539-547.
Robert-Seilaniantz, A., L. Navarro, R. Bari, and J.D. Jones. 2007. Pathological hormone imbalances. Curr. Opin. Plant
Biol. 10: 372-379. Ryu, C.M., A. Anand, L. Kang, and K.S Mysore. 2004.
Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 40: 322-331.
Scott, J.W., J.F. Wang, and P.M. Hanson. 2005. Breeding
tomatoes for resistance to bacterial wilt, a global view. In T. Momol, P. Ji, and J.B. Jones (eds.), Proceeding of 1st International Symposium on Tomato Diseases, Acta Hort.,
pp. 161-172.
Senthil-Kumar, M., G. Govind, L. Kang, K.S. Mysore, and M. Udayakumar. 2007a. Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-
|
|