426
Botanical Studies, Vol. 50, 2009
Province (China) and C. notoensis from Japan. No fossils of the genus have been recorded from western Europe or eastern North America.
Of the four incense cedars, the taxonomic status of C. formosana has been exceptionally controversial. It was first attributed to Libocedrus macrolepis when it was reported from Taiwan by Hayata (1908). Because of its smaller and obtuse lateral leaves (see Figure 1 in this study), 3-anthered stamens, and larger cones (pollen bearing cones 4-6 mm long and ovule bearing cones 12-15 mm long) and seeds (5 mm long), C. formosana was considered different from plants on the mainland by Florin (1930), thus it was recognized as L. formosana. Kudo (1 931 ), however, claimed that the characters used by Florin were variable and not sufficiently significant to recognize the Taiwanese plants as a distinct species. He therefore treated it as L. macrolepis var. formosana (Florin) Kudo. At that time Calocedrus was included in Libocedrus, which is now limited to species native to New Zealand and New Caledonia. After the recognition of Calocedrus by Li (1953), Florin (1956)
made the combination, C. formosana (Florin) Florin, for the Taiwanese incense cedar. Interestingly, western taxonomists preferred to treat C. formosana as a distinct species (e.g. Florin, 1956; Krussmann, 1985; Kvacek, 1999) while eastern taxonomists (e.g. Li and Keng, 1994) followed Kudo. Hence, a new approach to resolving these conflicting views is needed.
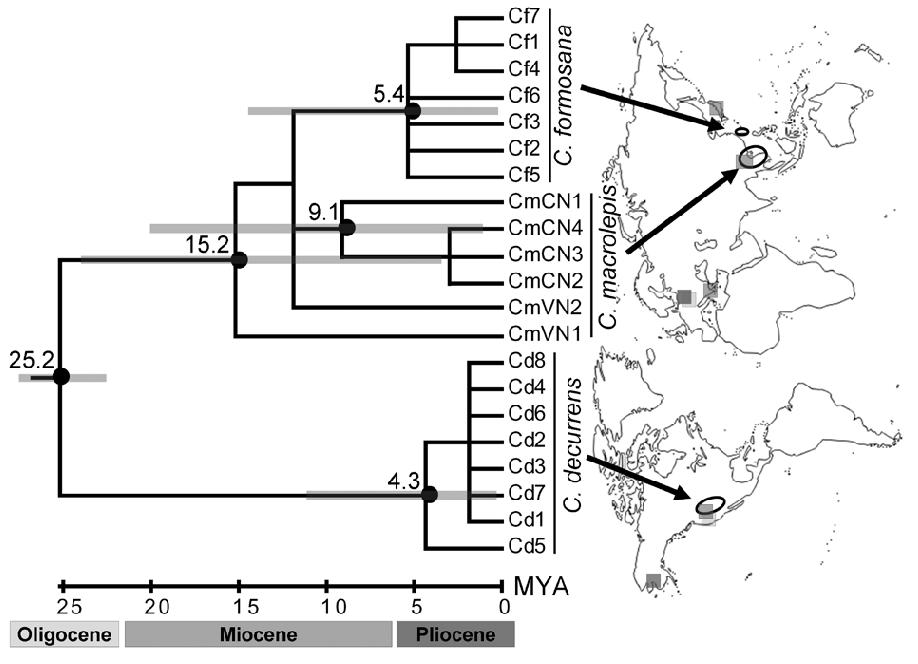
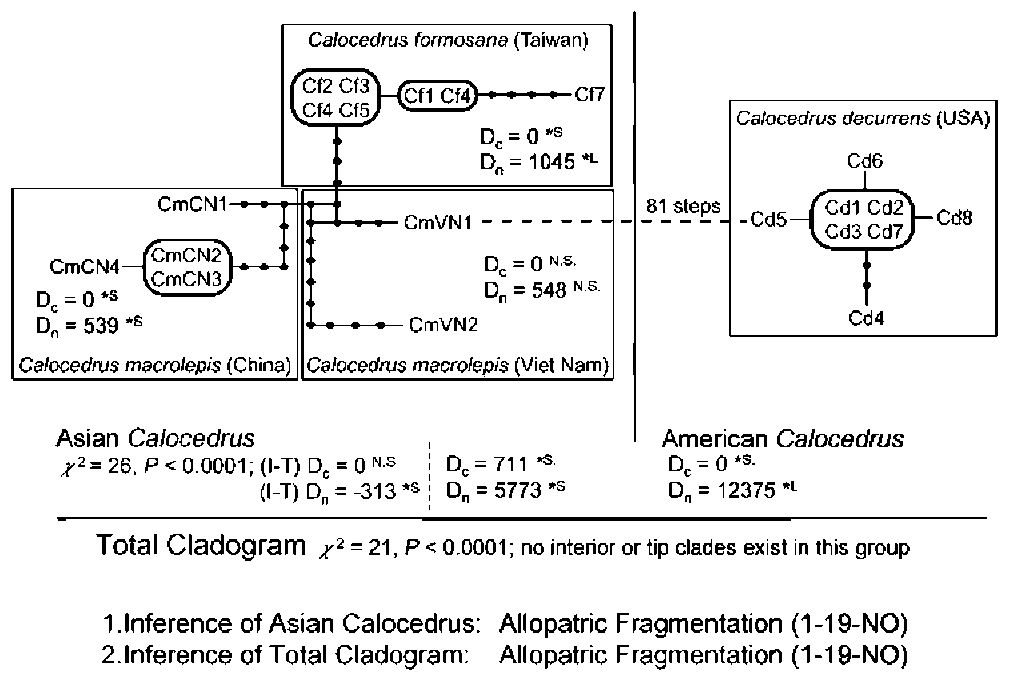
Sequence data from the internal transcribed spacers of nuclear ribosomal DNA (nrITS) have been utilized successfully to elucidate phylogenetic relationships at the generic and specific levels of both gymnosperms (e.g. Liston et al., 1996; Cheng et al., 2000) and angiosperms (e.g. Shi et al., 1998; Wen and Shi, 1999). Here we present our comparative analyses of nrITS from 21 individuals of three taxa of Calocedrus, C. decurrens, C. formosana, and C. macrolepis. Relationships within the genus were inferred and discussed based on the sequence variation and reconstructed phylogenetic trees. Furthermore, phylogeographic inferences and fossil records were used to determine the historical scenarios that might account for the present disjunct distribution of Calocedrus.
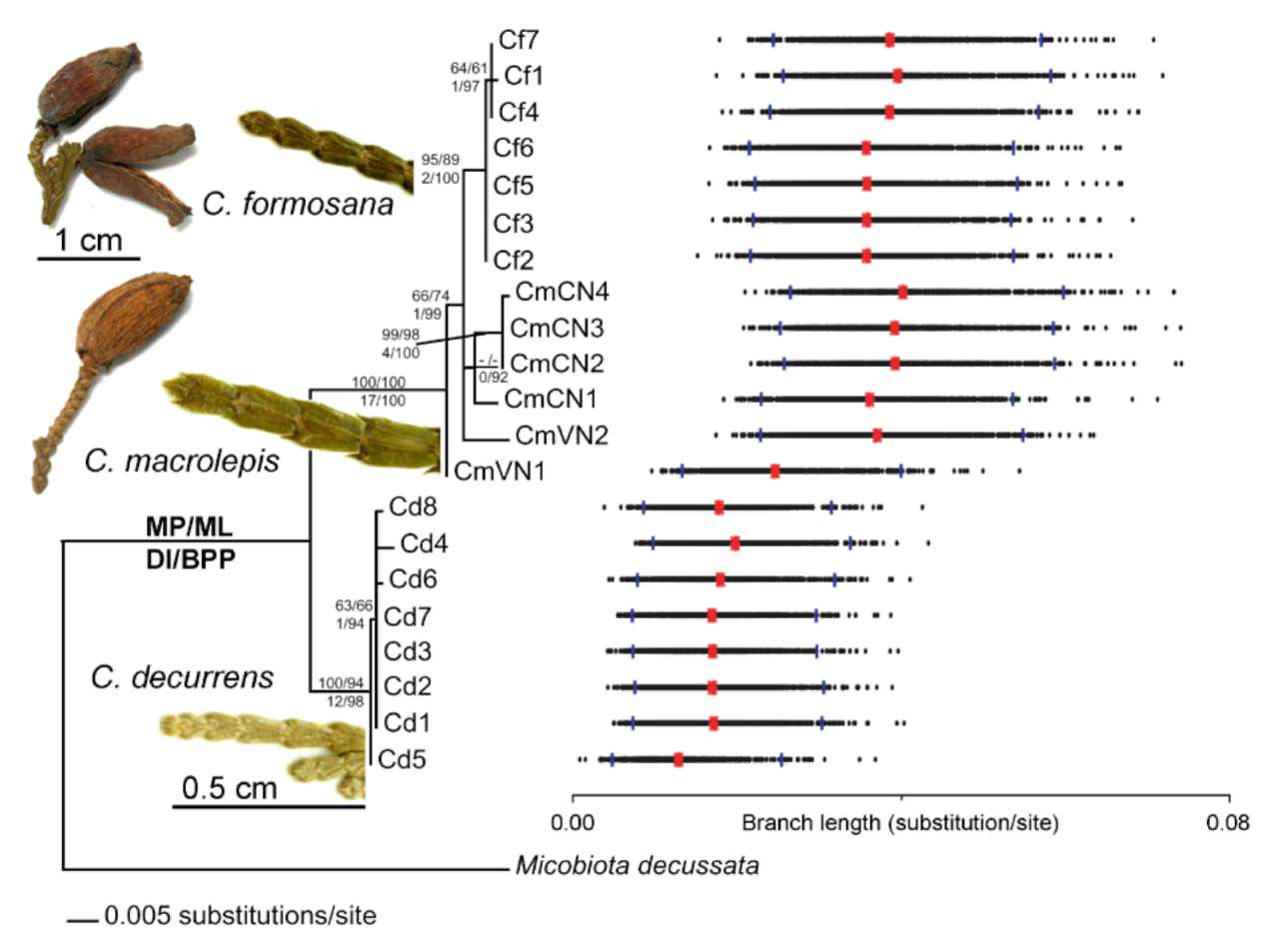
Figure 1. ML phylogeny for Calocedrus (-lnL = 2379.00157; model: TrN + r). Branch support was calculated using MP bootstrapping (above branch before slash), ML bootstrapping (above branch after slash), Decay index (below branch before slash), and BPP (below branch after slash). Bayesian relative rate (BRR) test is shown next to individual codes; orange points indicate MLEs; and blue lines at both edges illustrate 95% BPPs. Estimated values overlap 95% BPPs, indicating a constant evolutionary rate. Left figures illustrate leaf branches for three species of Calocedrus (scale bar = 0.5 cm) and ovulate cones for C. formosana and C. macrolepis (scale bar = 1 cm); C. formosana and C. macrolepis (HAST accessions 65530 and 43827, respectively).