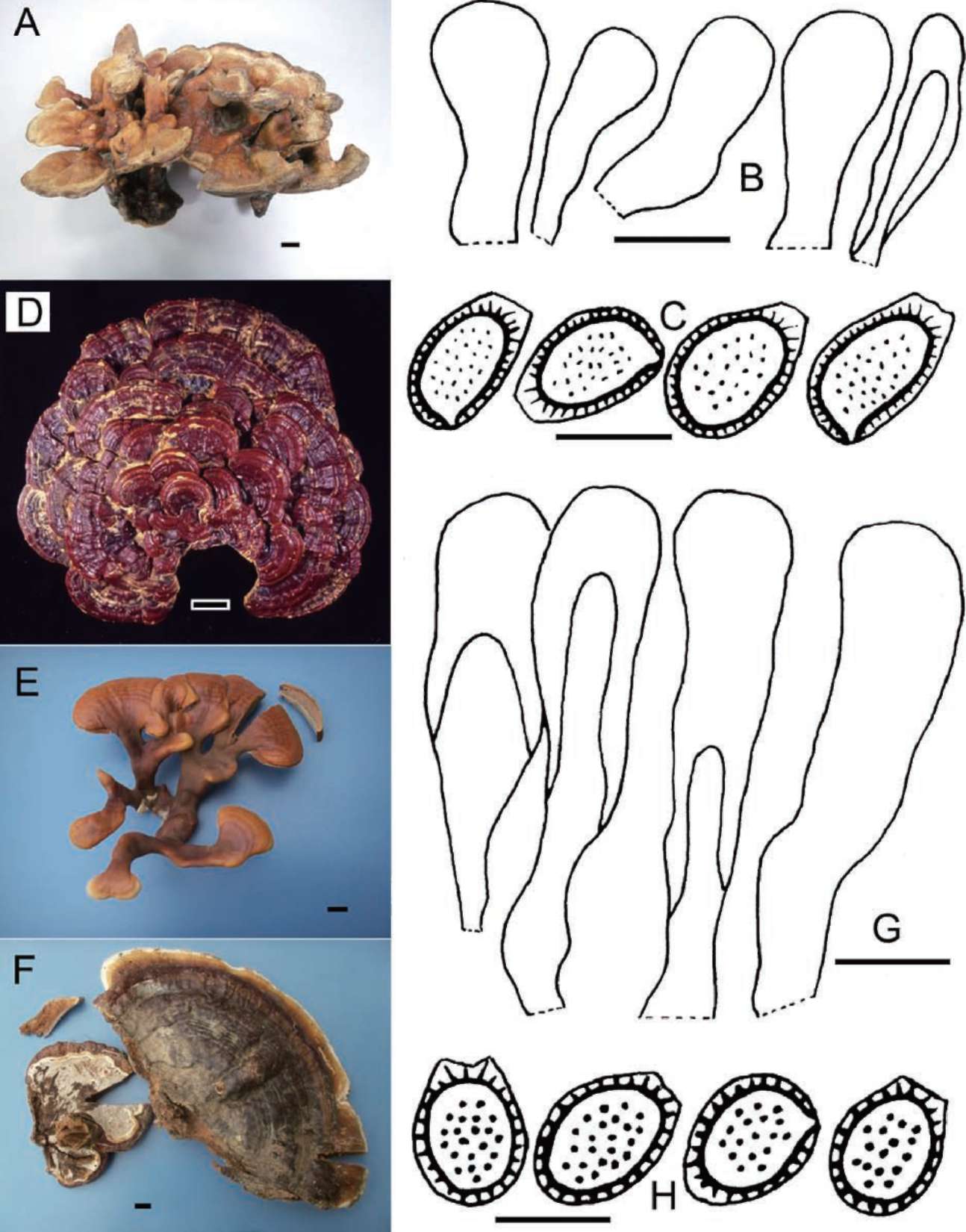
pileum, Dr. Tsuyoshi Hosoya (TNS) for the loan of a specimen of ' G. lucidum' collected from Chiayi, Taiwan and Dr. Tsutomu Hattori (TFM) for arranging the loan of two specimens of ' G. lucidum' collected from Ibaraki, Japan. We thank Ms. Shih-Yi Yu for her assistance in obtaining DNA sequence data for this study. This project was supported by the National Museum of Natural Science (ROC), Foundation of the National Museum of Natural Science (ROC), and a Postdoctoral Fellowship Grant of the National Science Council (ROC), no. NSC96-2816-B-178-001,
to the senior author. The senior author also appreciates the support of a project grant (30270006) from the National Nature Science Foundation of China for her initial Gano-derma work in mainland China through Dr. Y.-J. Yao.
LITERATURE CITED
Bazzalo, M.E. and J.E. Wright. 1982. Survey of the Argentine species of the Ganoderma lucidum complex. Mycotaxon 16: 295-325.
Chang, T.T. 1983. Studies on biology of several species of Ganoderma in Taiwan [Master's thesis]. National Taiwan University, Taipei, 130 pp.
Chang, T.T. and T. Chen. 1986. Studies on nuclear behavior, mating type and heterokaryosis of several species of Gano-derma in Taiwan. Plant Prot. Bull. (Taiwan, R.O.C.) 28:
231-240.
Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791.
Gilbertson, R.L. and L. Ryvarden. 1986. North American Polypores 1. Fungiflora, Oslo, Norway, 433 pp.
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucl. Acids Symp. Ser. 41: 95-98.
Hseu, R.S., H.H. Wang, H.F. Wang, and J.M. Moncalvo. 1996.
Differentiation and grouping of isolates of the Ganoderma lucidum complex by random amplified polymorphic DNA-PCR compared with grouping on the basis of internal transcribed spacer sequences. Appl. Environ. Microbiol. 62:
1354-1363.
Hou, D. 1950. A new species of Ganoderma from Taiwan. Q. J.
Taiwan Mus. 3: 101-105.
McKenzie, E.H.C. and M.N. Foggo. 1989. Fungi of New Zealand subantarctic islands. NZ J. Bot. 27: 91-100.
Moncalvo, J.M. and L. Ryvarden. 1997. A nomenclatural study of the Ganodermataceae Donk. Fungiflora, Oslo, Norway,
114 pp.
Moncalvo, J.M., H.F. Wang, and R.S. Hseu. 1995a. Gene
phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycol. Res. 99: 1489-1499.
Moncalvo, J.M., H.F. Wang, H.H. Wang, and R.S. Hseu. 1995b.
The use of ribosomal DNA sequence data for species identification and phylogeny in the Ganodermataceae. In P.K. Buchanan, R.-S. Hseu and J.-M. Moncalvo (eds.), Gano-