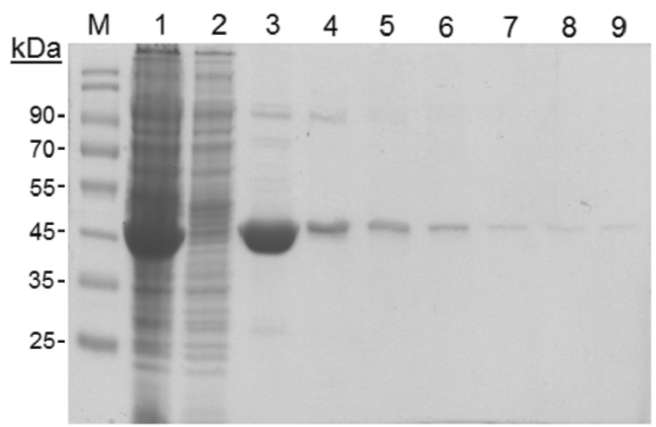
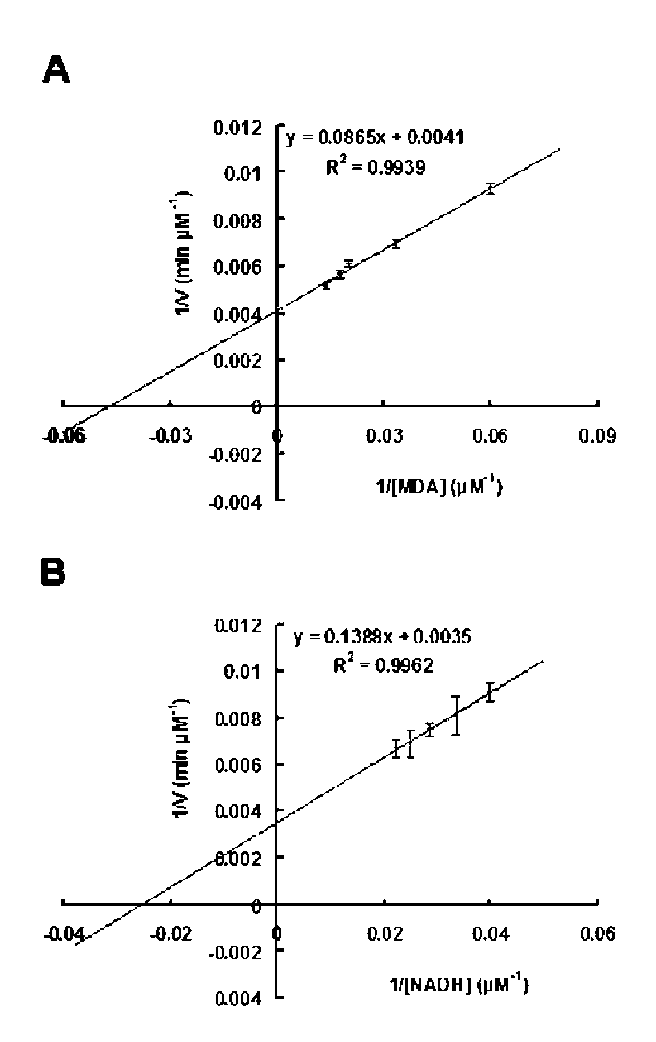
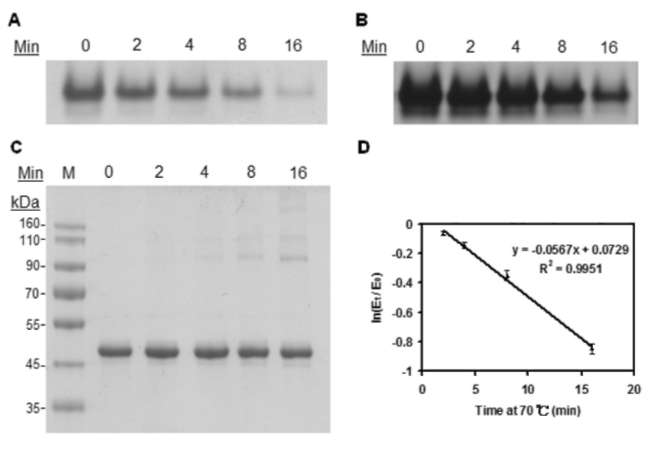
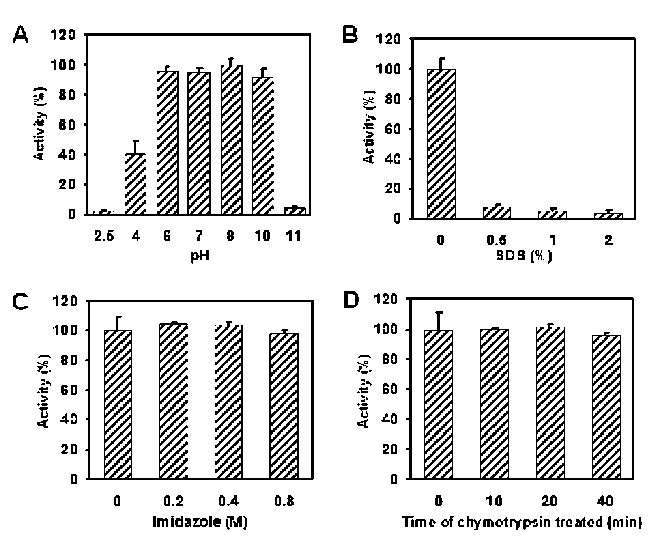
40 Botanical Studies, Vol. 51, 2010
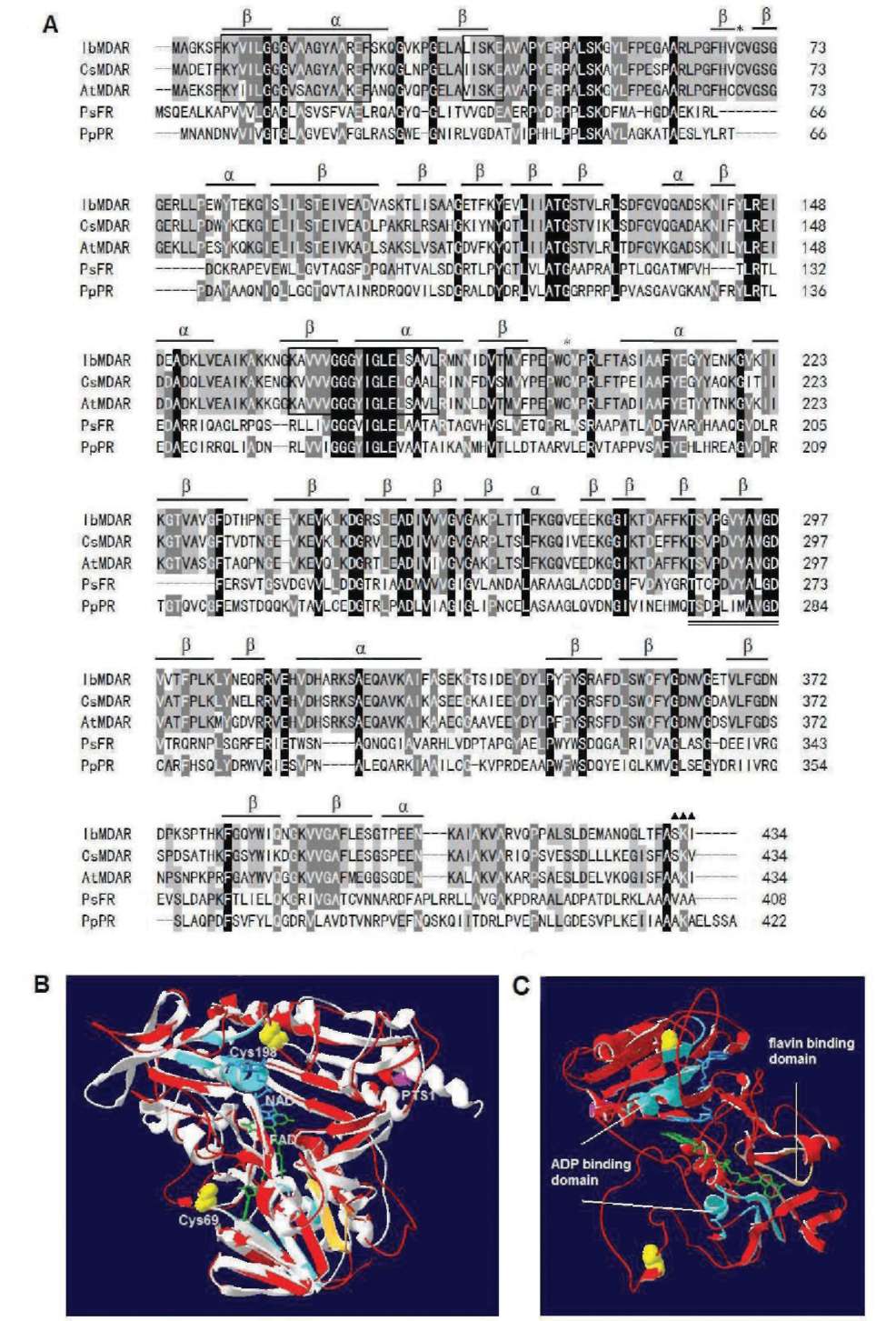
Figure 1. Alignment of the amino acid sequences of MDAR with other organisms and 3-D homology structure. (A) Sequence alignment: IbMDAR (Ipomoea batatas, this study), CsMDAR (Cucumis sativus, BAA05408), AtMDAR (Arabidopsis thaliana, Q9LFA3), PsFR (Pseudomonas sp. KKS102 ferredoxin reductase, Q52437), and PpPR (Pseudomonas putida putidaredoxin reductase, P16640). Identical amino acids in all sequences are shaded black, conservative replacements are shaded gray. Protein secondary structure was predicted by SWISS-MODEL program and represented as a helices and p strands. Stars denote Cys69 and Cys198, triangles denote putative PTS1 (peroxisomal targeting signal) sequence, amino acid domains involved in binding of the ADP moiety of FAD and NAD(P)H are boxed, a conserved 11-amino acid domain (positions 287-297) important in binding of the flavin moiety of FAD is double underlined; (B) 3-D homology structure of MDAR. The structural model of MDAR was created based on the known structure of putidaredoxin reductase (PDB code 1q1r) from Pseudomonas putida via SWISS-MODEL program and was superimposed to obtain better structure alignment and DeepView Swiss-PdbViewer v3.7 programs. Superimposition of MDAR (red) and putidaredoxin reductase (PDB code 1q1r) from Pseudomonas putida (white gray) was shown by using protein solid ribbons. Yellow balls denote Cys69 and Cys198, pink ribbon denotes PTS1 sequence, green denotes FAD, blue denotes NAD; (C) Side view of the same structure. Light blue ribbon denote ADP moiety of FAD and NAD(P)H binding domain, light orange ribbon denotes flavin moiety of FAD binding domain.