YU et al. ― Phylogenetic relationships of Antrodia
57
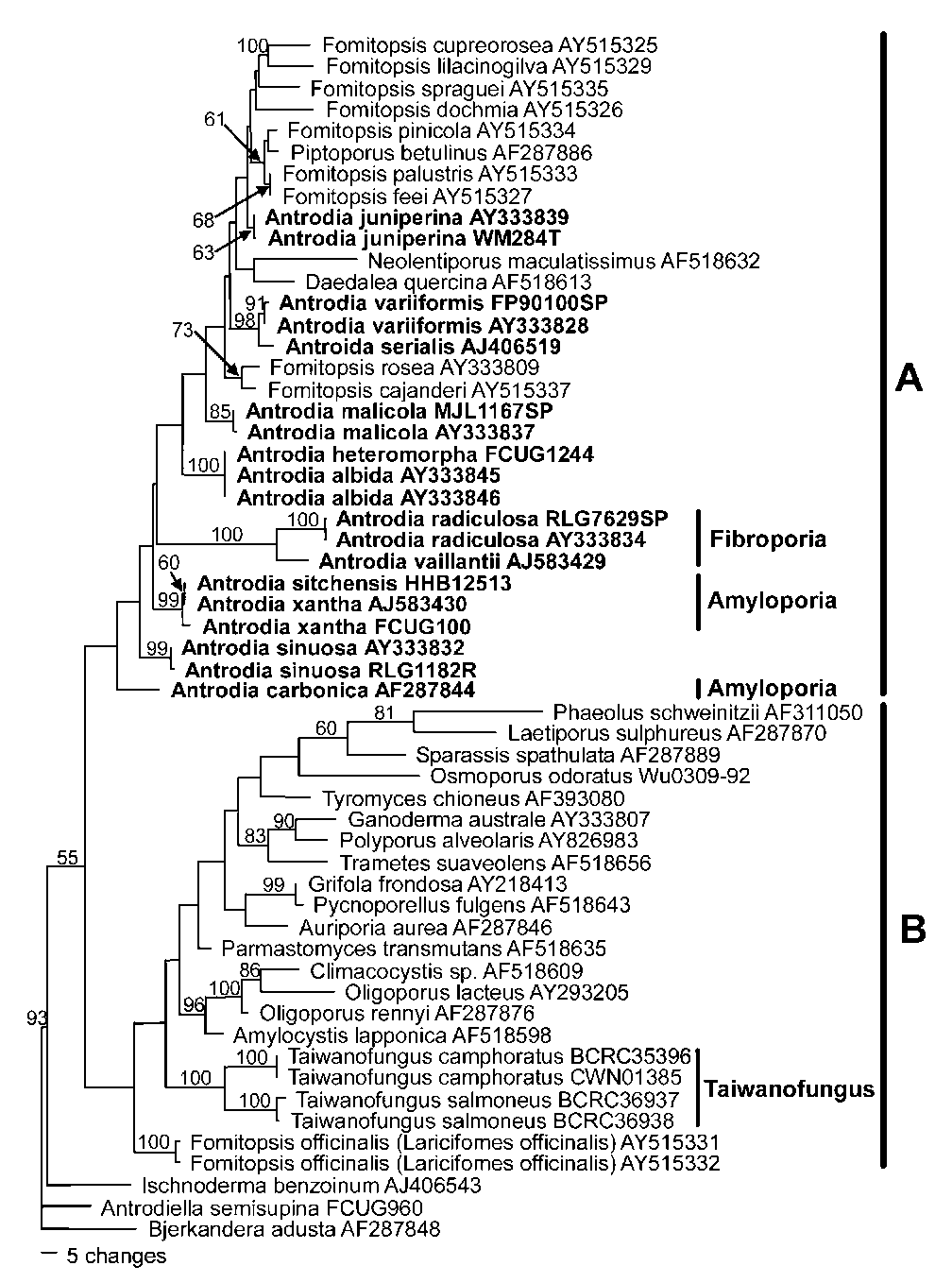
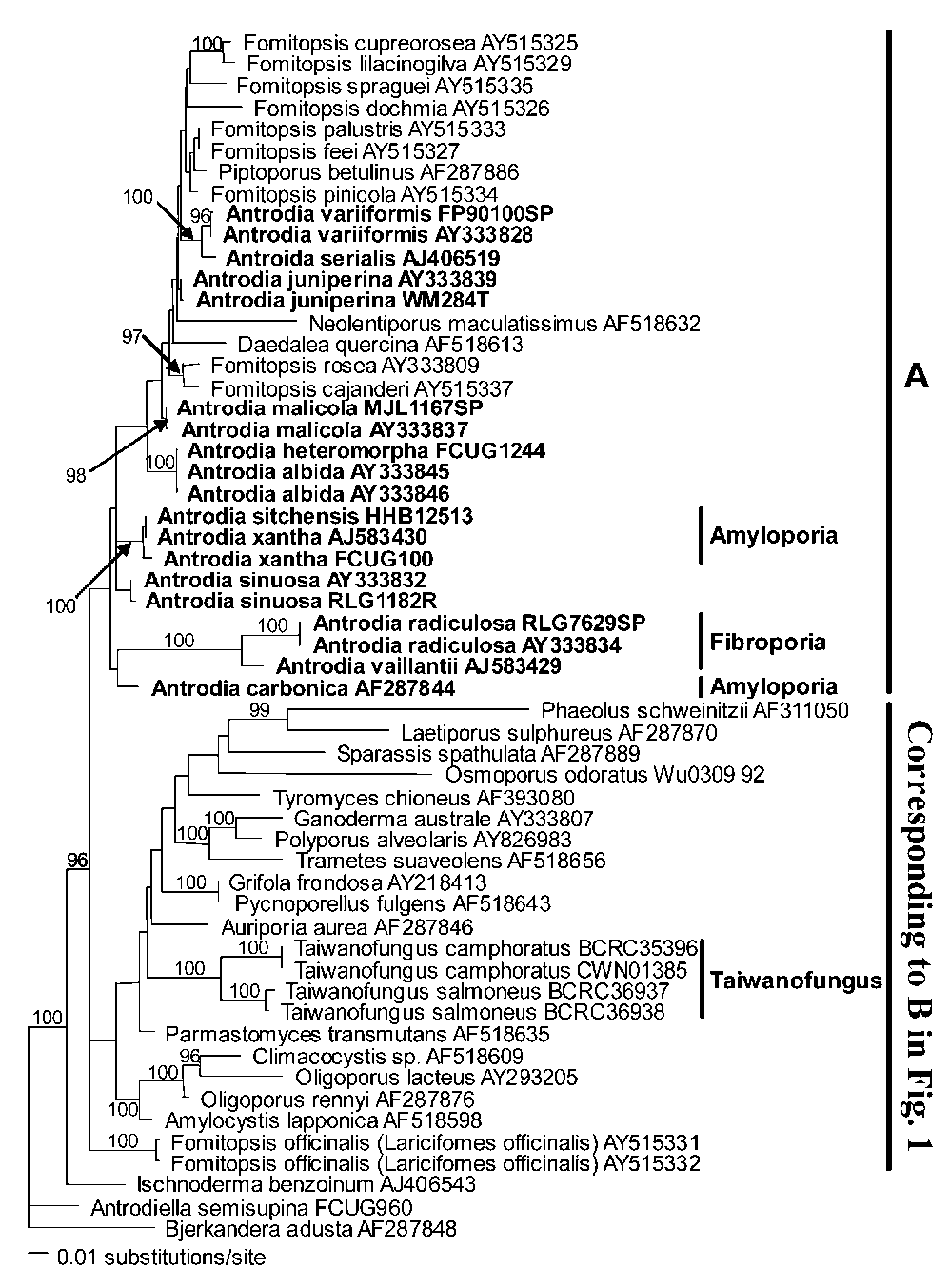
Figure 1. One of the ten most parsimonious trees derived from partial nuc-LSU rDNA sequence data. Bootstrap values are shown at nodes supported by no less than 50% from 1000 replicates. TL = 911, CI = 0.425, RI = 0.645.
1 and 2). These two species differ from other species of Antrodia by having a fruiting body with a rhizomorphic margin, and a tetrapolar mating system (Lombard, 1990) while most species of Antrodia possess a bipolar mating system. Antrodia malicola is an exception, with a homothallic mating system. Although Fibroporia gossipina was not included in our study, this species formed a well-supported clade with Fibroporia vaillantii in a previous study (Kim et al., 2001). It is, therefore, evident that the three members of Fibroporia, Fib. radiculosa, Fib. gossipina, and Fib. vaillantii, are closely related. Molecular results and sexuality along with morphological features support Fibroporia being a distinct genus.
Three species of Amyloporia with amyloid skeletal hyphae, i.e., Amy. carbonica, Amy. sitchensis, and Amy.
xantha, nested within clade A. However, only two of them, Amy. xantha (the type of Amyloporia) and Amy. sitchensis formed a very strongly supported subclade in clade A (Figures 1 and 2). Amyloporia carbonica is separate from this subclade, but its position remains unresolved (Figures 1 and 2). In addition to molecular data indicating that Amyloporia is not a monophyletic genus, morphological delimitation from Taiwanofungus also appears problematic.
Both genera have amyloid skeletal hyphae, but the two species of Taiwanofungus endemic to Taiwan, T. camphoratus and T. salmoneus, formed a well-supported subclade within clade B (Figures 1 and 2), well separated from Antrodia, Fibroporia, and Amyloporia. This means that the generic status of Amyloporia cannot be recognized.