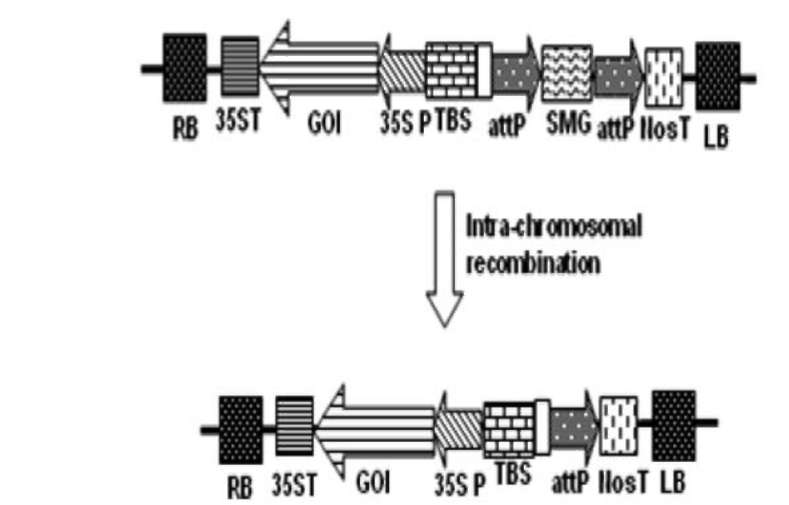
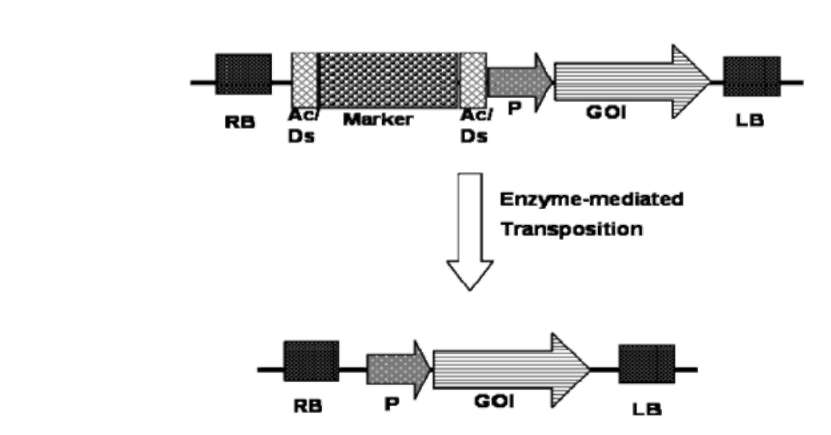
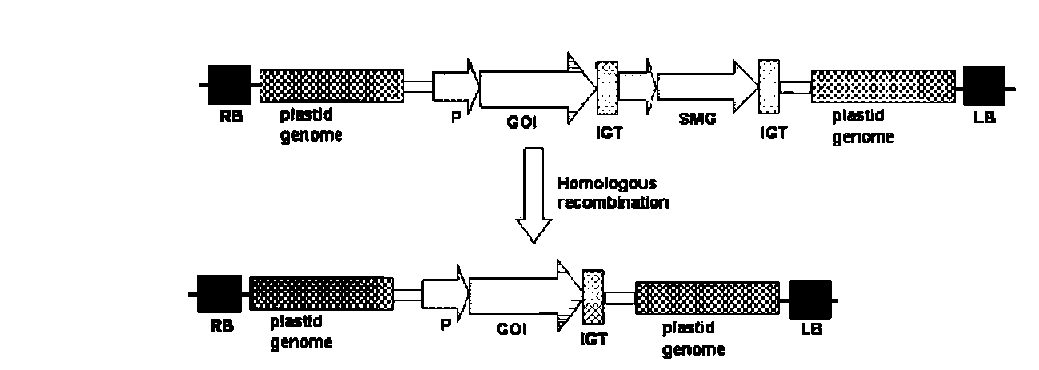
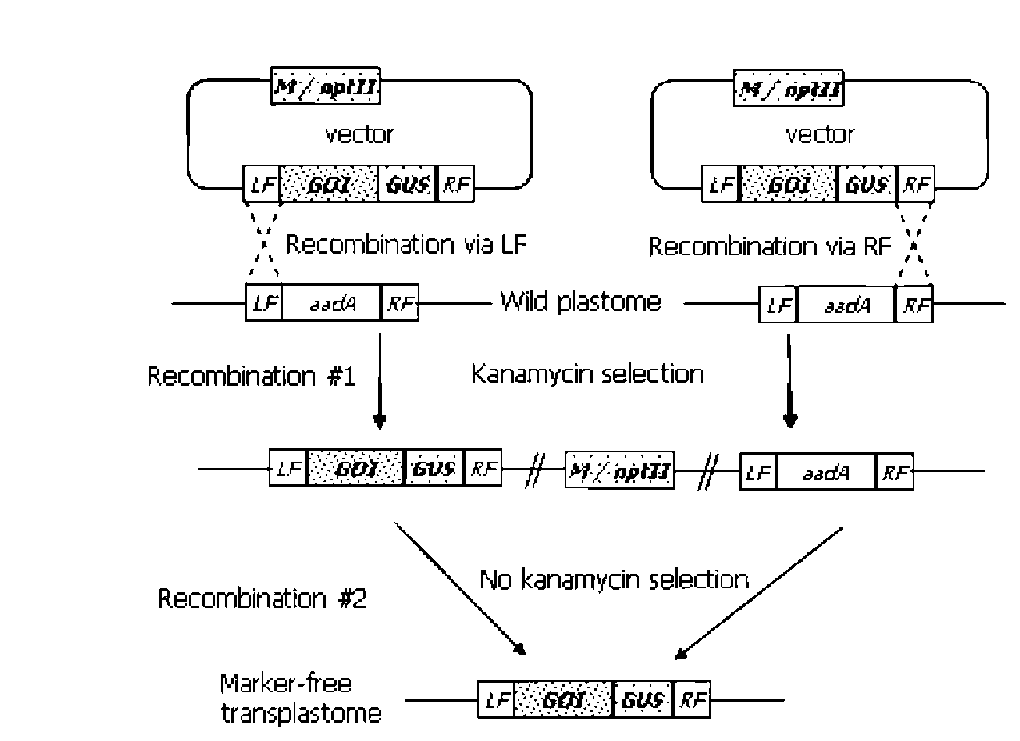
UPADHYAYA et al. — Strategies to develop marker-free transgenics 279
selective growth of the transformants, and the researcher has to screen several putative transformants to confirm the integration of the transgene(s), which is expensive and time consuming. Uncontrolled regeneration of a large number of chimeric plants is another drawback in the case of selection without the use of antibiotics as observed in
tobacco (Li et al., 2009).
Positive selection system
Some biochemical marker genes enable the identification and selection of transformed cells without injury or death of the non-transformed cells. In this method, the selection marker genes code for enzymes that give the transformed cells a capacity to metabolize specific nutrient substances not usually metabolized by normal plants (Figure 1). This fact will provide the transformed cells an advantage over the non-transformed ones. Addition of this substance in the culture medium, as nutrient source during the regeneration process, allows normal to enhanced growth and differentiation of transformed cells while non-transformed cells will not be able to grow and regenerate. Use of GUS, manA, and xylA genes are some of the popular positive selection systems.
The GUS gene. The ^-glucuronidase (GUS) gene isolated from E. coli (Jefferson et al., 1986) is to date the most widely used reporter gene in genetically modified plants. Agronomically, the E. coli GUS gene has no value to crop plants. In this system of positive selection, a glucuronide derivative of benzyladenine (benzylad-enine N-3-glucuronide) is used as selection agent. The glucuronide supplemented in the selection medium is hy-drolyzed by the GUS enzyme produced in the transformed cells, forming active cytokinin (benzyladenine). The released cytokinin acts as a stimulator for transformed cell regeneration, while non-transformed cell development is slow as the inactive form of the selective agent does not influence the non-transformed cells. This system was successfully used to effectively recover transgenic plants of tobacco (Joersbo and Okkels, 1996; Okkels et al., 1997).
The manA gene. Phosphomannose isomerase (PMI, EC 5.3.1.8) is an enzyme that converts mannose-6-phos-phate to fructose-6-phosphate, an intermediate of glycoly-sis that positively supports the growth of plant cells. This gene helps the transformed plant cells to utilize mannose as a carbon source to grow and differentiate on media containing mannose as a selection agent. In 1984, the gene coding for phosphomannose isomerase (manA or pmi) was first isolated from E. coli by Miles and Guest. However, its first application in plants as a selection marker gene was reported by Joersbo et al. (1998). Though most plant species are sensitive to mannose, a few species— especially dicots like carrot, tobacco, sweet potato and legumes—have shown a considerable insensitivity towards this sugar. Plants like sugar beet, maize, wheat, oat, barley, tomato, potato, sunflower, rapeseed, pea, onion, and rice are extremely sensitive and have been successfully transformed using pmi as a positive selection marker
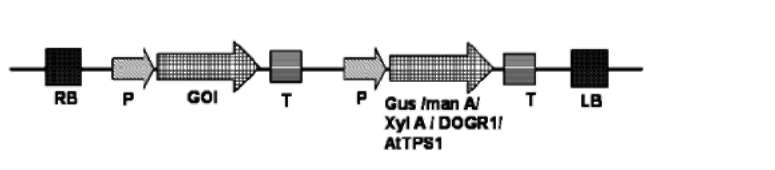
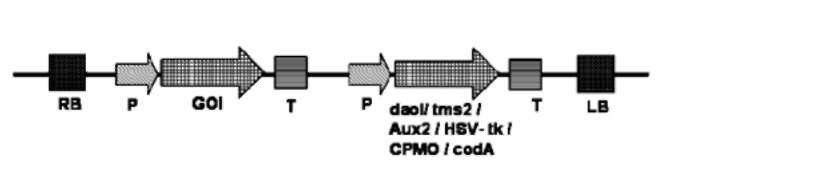
Figure 1. T-DNA of plant transformation vector harboring positive section marker genes, GUS, P-glucuronidase; pmi, phosphomannoisomerase; xylA, Xylose isomerase A; DOGR1, 2-deoxyglucose-6-phosphate phosphatase; AtTPS1, trehalose-6-phophate synthase; used for producing marker-free transgenic plants.
(Joersbo et al., 1998, 2000; Negrotto et al., 2000; Wang et al., 2000; Bhalla-Sarin et al., 2004; Gadaleta et al., 2006;
Ku et al., 2006; Aswath et al., 2006; Penna et al., 2008).
Some plant transformation protocols that use the positive selection with pmi were found to be at least 10 times more efficient than the traditional protocols based on kanamycin selection (Wright et al., 2001). Jain et al. (2007) reported a maximum of 56% efficiency for stable transformants using mannose selection in sugarcane (Saccharum spp.).
The xylA, DOGR1 and AtTPS1 genes. The xylA gene isolated from Thermoanaerobacterium thermosul-furogenes or Streptomyces rubiginosus (Haldrup et al., 1998a) encodes for enzyme xylose isomerase, which catalyzes the isomerization of D-xylose to D-xylulose. It also catalyzes the isomerization of glucose to fructose, and is thus also termed glucose isomerase. Transgenic cells expressing the xylA gene can utilize xylose as a sole carbohydrate source and proliferate while non-transgenic cells starve. Using this system, transgenic plants of potato, tobacco, and tomato were successfully selected in xylose-containing media (Haldrup et al., 1998b; 2001). However, a problem initially encountered when xylose was used in the selection medium was the induction of callus, which resembled the auxin effect. This problem can be solved by adding auxin efflux inhibitor, triiodobenzoic acid (TIBA) (Nissen and Minocha, 1993), an antiauxin, p-chlorophe-noxyisobutyric acid (PCIB) (Kasai and Bayer, 1995), or an ethylene inhibitor, aminoethoxyvinylglycine (AVG) (Hall et al., 1985), to the selection medium.
In another system, the DOGR1 gene isolated from yeast encoding 2-deoxyglucose-6-phosphate phosphatase (2-DOG-6-P) gives resistance to 2-deoxyglucose (2-DOG). In plant cells 2-DOG is converted into 2-DOG-6-phos-phate (2-DOG-6-P), which is a competitor of glucose-6-phosphate. When over-expressed in transgenic plants, it was used as a positive selection system for tobacco and potato plants (Kunze et al., 2001). Recently, a new positive selection marker gene AtTPS1, encoding trehalose-6-phophate synthase, has been developed (Leyman et al., 2008). In this system, the selection agent used is the nontoxic and common sugar glucose. Wild-type non-transformed Arabidopsis thaliana plantlets germinated on glucose had small white cotyledons and remained petite due to stoppage of the photosynthetic mechanism by external sugar while transgenic plants expressing AtTPS1 became insensitive to glucose. The selectable marker gene