304
Botanical Studies, Vol. 51, 2010
electroporation procedure for mycelial cell transformation was developed. The heterologous expression of the enhanced green fluorescent protein (egfp) in F. velutipes is reported. We also demonstrate that egfp was expressed in mature fruiting bodies and remained stable in monokaryons produced from meiosis.
MATERIALS AND METHODS
Strains and media
Flammulina velutipes BCRC 37086 was purchased from the Bioresources Collection and Research Center (Hsinchu, Taiwan) and grown in either PDA (Potato dextrose agar, Difco, Detroit, MI, USA) or PDB (Potato dextrose broth, Difco) at 25°C. The Escherichia coli DH5a (GIBCOBRL, Life Technologies, Grand Island, NY, USA) was used for DNA manipulation and grown in LB medium (Sigma Chem. Co., St. Louis, MO, USA) at 37°C.
Fruiting body development of F. velutipes
A medium composed of 65% sawdust and 35% rice bran was placed into a 500-ml flask and autoclaved for 1 h. Capped flasks inoculated with mycelial plugs were incubated at 25°C in the dark for 3 weeks. After vegetative mycelia were growing all through the medium, fruiting was induced by water addition, a temperature shift from 25°C to 10°C, and light exposure. To achieve the appearance of primordia and the maturation of fruiting bodies, the flasks were incubated under the induction conditions until mature fruiting bodies appeared.
Plasmid construction
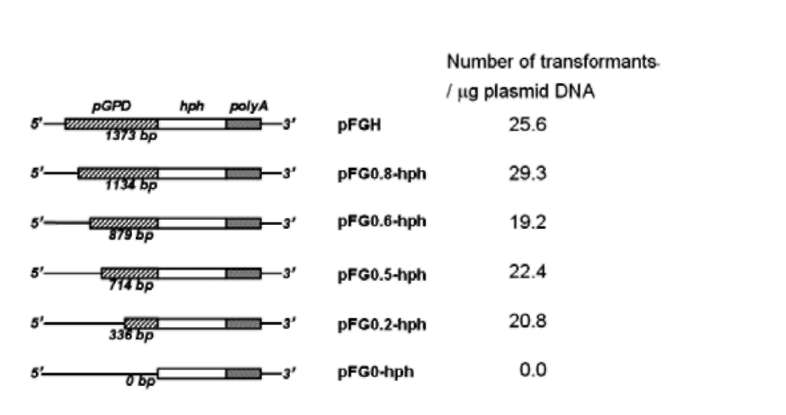
The plasmid pFGH was used as a backbone (Kuo et al., 2004). Primers used to amplify reporter genes and gpd promoter regions (pGPD) for deletion analysis are shown in Table 1. Promoters were amplified from F. velutipes genomic DNA; egfp gene were amplified from pHygEGFP (BD Bioscience, Palo Alto, CA). The resulting plasmids (Figure 1) were used for the transformation experiments.
Transformation procedure
Transformation was modified as in a previous paper (Kuo et al., 2004). A four-day-old liquid culture of mycelia was blended with a Waring blender and then incubated overnight with gentle shaking at 25°C. Mycelial fragments were collected by centrifugation at 3,000 g, followed by washing with P buffer (0.02 M phosphate buffer, pH 5.8, 0.6 M mannitol) and treatment with 2 mg/ ml Lysing enzymes (Sigma) for 3 h. After washing free of enzyme, 0.5 g (wet weight) mycelial fragments were mixed with plasmid DNA and subjected to electroporation. Electroporation was performed by BTX ECM 630 and 0.2-cm cuvettes (BTX, San Diego, CA, USA) with the electric pulse delivery setting of 25 fiF in capacitor, 100 Q in resistor, and 12.5 kV/cm in field strength. Transformants were selected on PDA plates containing 30 [ig/ml hygromycin (Sigma).
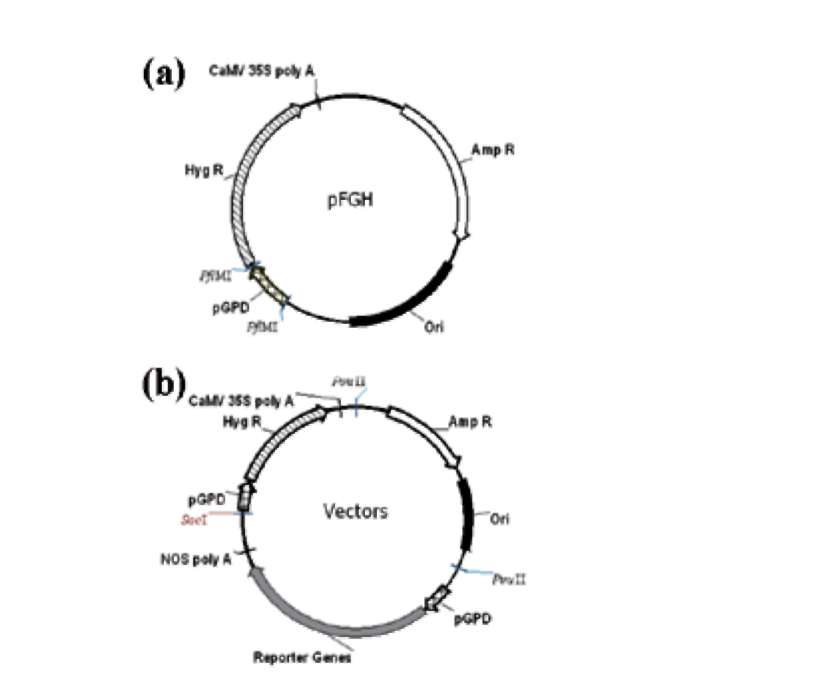
Figure 1. Organization of plasmids transformed. (a) Plasmids used in deletion analysis consisted of a pFGH vector backbone (Kuo et al., 2004). Different lengths of pGPD fragments were introduced by restriction enzyme Pfl MI; (b) Plasmid for the expression of egfp as reporter gene. The hygromycin resistance gene (Hyg R) and reporter gene were joined to the gpd promoter (pGPD). NOS poly A: nopaline synthase poly A signal. CaM-V35S poly A: CaMV 35S poly A signal. Amp R: the ampicillin resistance gene.
Detection of EGFP in transformants
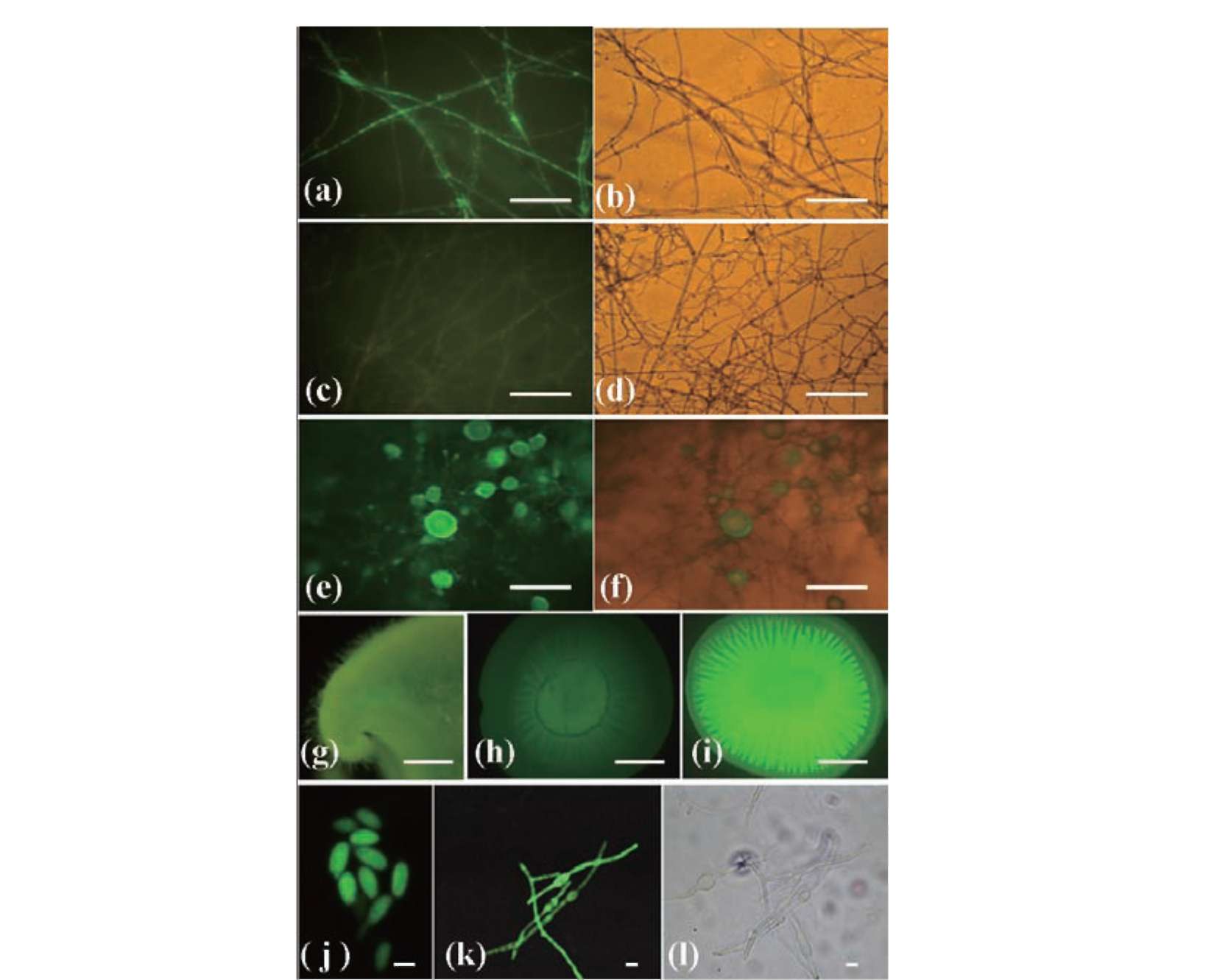
EGFP transformants were screened by a fluorescent microscope (E600, Nikon, Tokyo, Japan) using a Nikon B-2A filter (excitation filter, 450-490 nm; dichroic filter, 505 nm; barrier filter, 520 nm). Fruiting bodies of transformants and wild-type were observed using a stereo fluorescence microscope (SV11 APO/AxioCam MRc5, Carl Zeiss, Inc., Thornwood, NJ), and the fluorescent microphotographs were captured under the same exposure time.
Southern hybridization
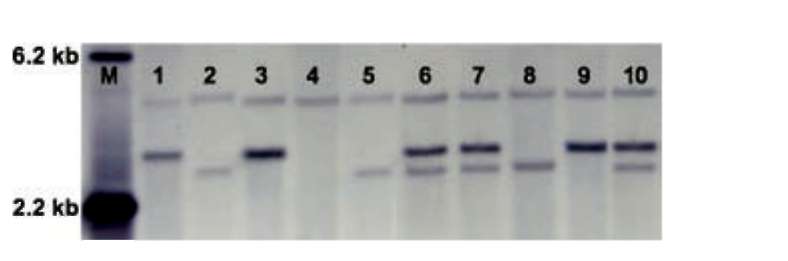
Genomic DNA isolation and the southern hybridization procedure were described previously (Kuo et al., 2004). Labeling of the DNA probe, hybridization, and signal detection were conducted by means of a Roche DIG-probe synthesis and detection kit (Roche, Mannheim, Germany) according to the manufacturer's instructions.
Western hybridization
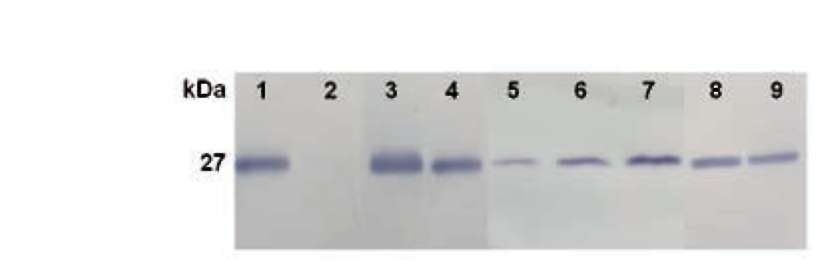
For western analysis of EGFP, F. velutipes transformants and the wild-type strain were grown for 7 days in PDB. Mycelia were collected and subsequently ground in liquid nitrogen with a mortar and pestle. A total of 50 mg mycelial powder was mixed with 1 ml protein extraction buffer (50 mM sodium phosphate, pH 7.4, 1 mM PMSF, 0.1% Triton X-100, 0.5 M NaCl) on