KO et al. ― Aquaperonospora taiwanesis gen. et sp. nov.
345
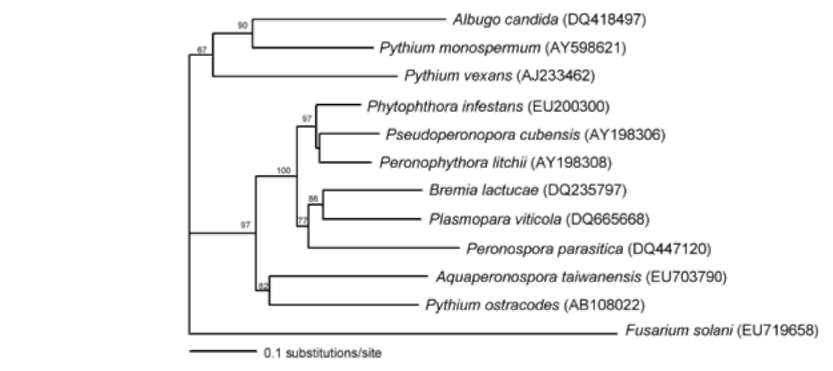
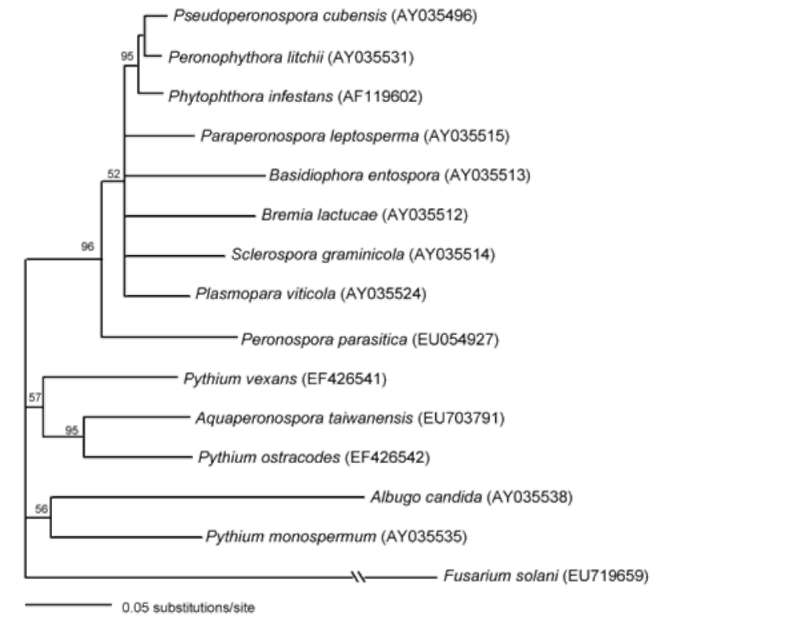
sequences was done using Kimura's two-parameter method (Kimura, 1980) of the program PAUP* 4.0 (Swofford, 1998). Bootstrap values were generated with 1000 replicate heuristic searches to estimate support for the clade stability of the consensus tree (Felsenstein, 1985).
TAXONOMY
Aquaperonospora Ko gen. nov.
Sporangiophora e myceliis, rigida, erecta, dichotome ramosa. Sporangia super ramunculis sporangiophorum simul formata, sporangiis additis super sporangiophoris e basi sporangiorum priorum interne vel externe prolificantibus factis successive. Zoosporae intra vesiculam externam sporangiorum formatae. Periplasma oogonii inconspicuum.
Etymology. Refers to its water habitat and sporangiophore morphology similar to that of the genus Peronospora.
Type species. A. taiwanensis Ko.
Aquaperonospora taiwanensis Ko sp. nov.
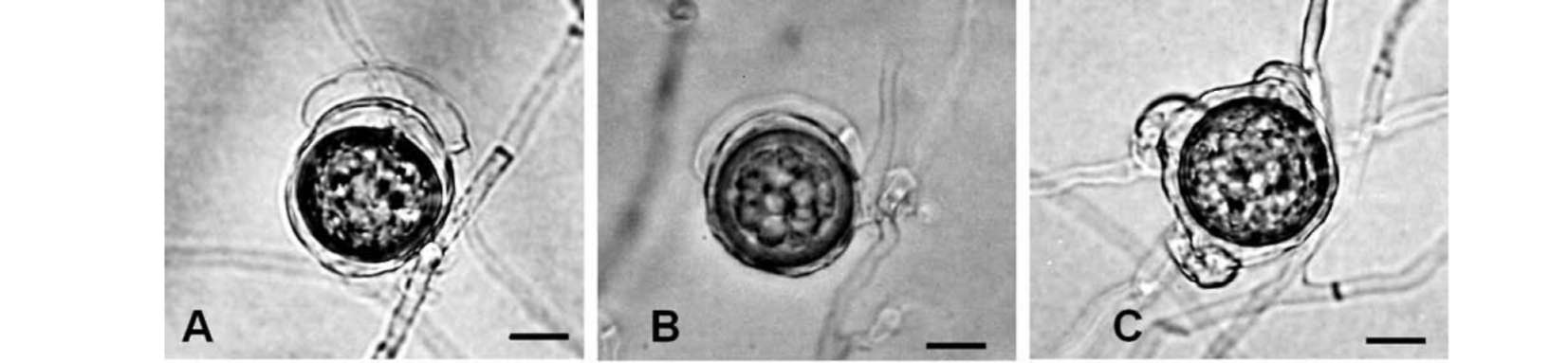
Hyphae coenocyticae, 3.0-4.4 μm latae. Sporangiophora e myceliis, rigida, erecta, dichotome ramosa, 475-1540 μm longa, 4.0-6.0 μm lata, unoquoque sporangia 1-30 ferenti. Sporangia citriformia, 33.0-41.0 x 22.0-29.6 μm, super ramunculis sporangiophororum simul formata, sporangiis additis super sporangiophoris e basi sporangiorum priorum interne vel externe prolificantibus factis successive. Zoosporae reniformes, 14-18 x 12-15 μm, intra vesiculam externam sporangiorum formatae. Periplasma oogonii inconspicuum. Oogonia globosa, terminalia vel intercalaria, laevia, 15-32 μm diam. Antheridia elongata, 12-21 x 3-5 μm, ad oogonium longitudine tota maximam partem singulariter adhaerentia. Oosporae apleroticae, 13-28 μm diam, pariete usque ad 3 μm crasso.
Etymology. Taiwanensis refers to the country of origin.
Holotype. HAST 109384 (dried culture), Herbarium,
Biodiversity Research Center, Academia Sinica, Taipei, Taiwan.
A culture from the holotype has been deposited at the Bioresource Collection and Research Center, Food Industry Research and Development Institute, Hsinchu, Taiwan (BCRC 34009).
Aquaperonospora taiwanensis was isolated by baiting from an irrigation ditch, with ten isolates collected in 2003 and two isolates in 2008. It is a fast growing organism compared to other fungi, with mycelia extending 23 mm/ day at 24°C on V-8 agar. The optimum temperature for growth was 32°C while the maximum and minimum temperatures were 40 and 12°C, respectively. The growth rate at 32°C was 32.3 mm for 24 h. Aquaperonospora taiwanensis did not produce asexual propagules on V-8 agar at 24°C under light. However, when a culture block was immersed in water and incubated at 24°C under light
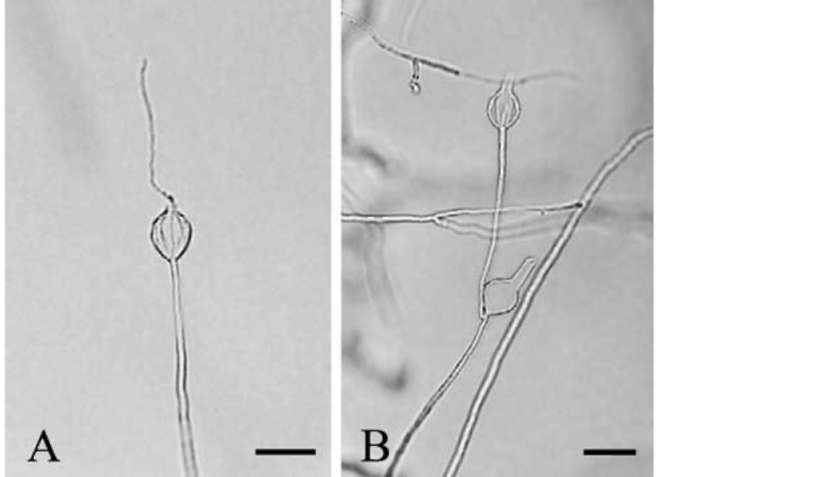
as described above, the organism produced numerous well differentiated Peronospora-like sporangiophores (Figure 1) within 24 h. Sporangiophores (475-1540 μm long, 4-6 μm wide) were rigid, erect, and dichotomously branched, bearing 1-30 sporangia each. Sporangia (33.0-41.0 x 22.0-29.6 μm) were lemon-shaped and formed synchronously on terminal branchlets of sporangiophores (Figure 2). Additional sporangia were mostly formed
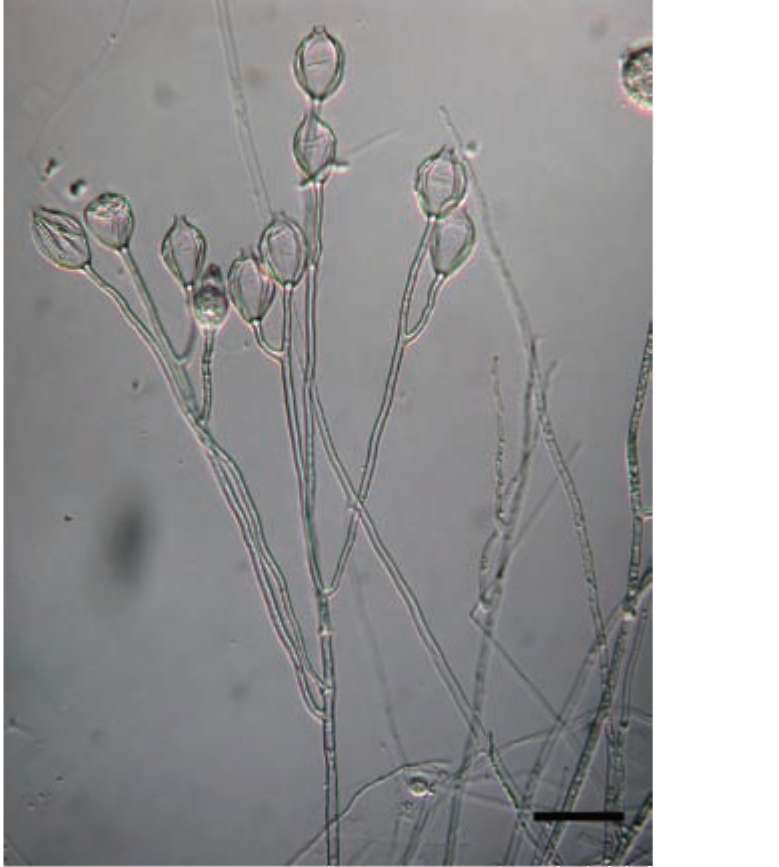
Figure 1. Differentiated sporangiophore of Aquaperonospora taiwanensis. Bar=50 [im.

Figure 2. Sporangiophores of Aquaperonospora taiwanensis. A, synchronous formation of sporangia on branchlets of a sporangiophore. Bar= 100 μm; B, concomitant discharge of cytoplasmic material from sporangia on a sporangiophore into vesicles during zoospore formation. Bar= 100 μm.