Botanical Studies (2010) 51: 413-420.

Molecular evaluation of interspecific hybrids between
Acer albopurpurascens and A. buergerianum var.
formosanum
Acer albopurpurascens and A. buergerianum var.
formosanum
Pei-Chun LIAO1,6, Huei-Chuan SHIH2,6, Tsair-Bor YEN3,6, Sheng-You LU4, Yu-Pin CHENG4 *, and
Yu-Chung CHIANG5 *
Yu-Chung CHIANG5 *
1 Department of Life Science, National Pingtung University of Science and Technology, Pingtung 912, Taiwan
2Department of Nursing, Meiho University, Pingtung 912, Taiwan
2Department of Nursing, Meiho University, Pingtung 912, Taiwan
3Department of Tropical Agriculture and International Cooperation, National Pingtung University of Science and Technology, Pingtung 912, Taiwan
4Division of Botanical Garden, Taiwan Forestry Research Institute, Taipei 10066, Taiwan
5Department of Biological Sciences, National Sun Yat-Sen University, Kaohsiung 80424, Taiwan
5Department of Biological Sciences, National Sun Yat-Sen University, Kaohsiung 80424, Taiwan
(Received June 8, 2009; Accepted January 20, 2010)
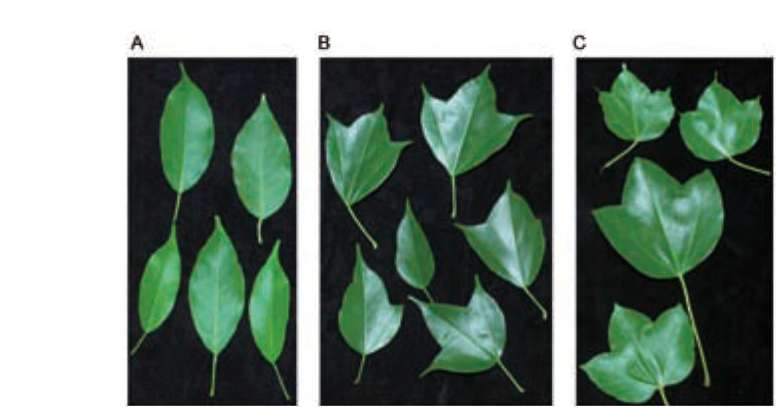
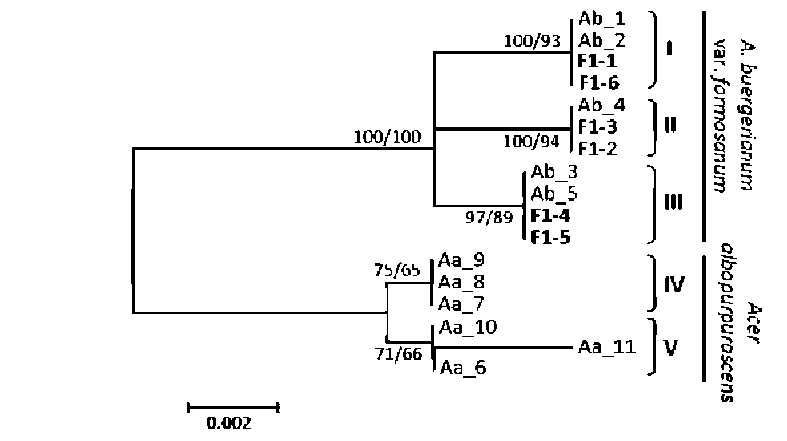
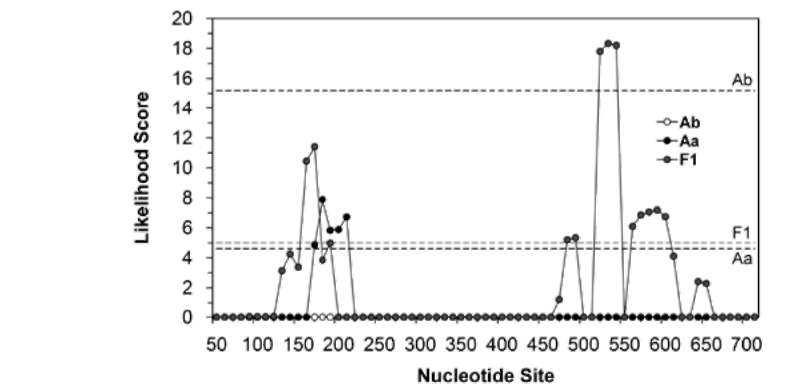
ABSTRACT. Ex situ conservation is a common strategy for the restoration of endangered species. Botanical gardens play an important role in the restoration of plants; however, hybridization could happen if transplantation is not considered carefully. Two naturally allopatric taxa Acer albopurpurascens and A. buergerianum var. formosanum, grown in the Taipei Botanical Garden, hybridized spontaneously. Hybridization was shown by the presence of intermediate phenotypes and genetic data. To confirm the direction of pollen flow, indirect evidence of maternally inherited chloroplast DNA indicated that A. buergerianum var. formosanum was the pollen receiver. The hybrids shared parts of ancestral genetic variations with their parents. Furthermore, several novel haplotypes that were slightly different from their parents were cloned, which revealed higher genetic diversity of the hybrid population than the parental populations. Such a phenomenon is called the rare allele phenomenon. In comparison with haplotypes of parents and hybrids, and according to the estimation of minimum recombination events and the likelihood ratio tests, the genetic variations were brought about by recombination events, such that the rare allele phenomenon might be related to the recombination. The hybridization that occurred in the botanical garden underscores the importance of spatial isolation when carrying out ex situ plantations.
Keywords: Acer; Ex situ conservation; Hybridization; Rare allele phenomenon; Recombination.
INTRODUCTION
Hybridization is commonly observed between closely related species (e.g. Kirk et al., 2004; Edwards et al., 2006) and even between distantly related species (e.g. Dunn and Lindstrom, 2008). In fact, hybridization may be viewed as an "invasion of the genome" from a genetic viewpoint (Mallet, 2005), and hybridization is potentially of evolutionary significance in the formation of species complexes (Rieseberg, 1995). The invasion of the genome promotes genetic evolution, and the rearrangement of parental species genomes may widen the adaptive range
for survival (heterosis). Through this process, life becomes more adaptive and diversified, especially in plants (Erickson and Fenster, 2006; Smith and Baum, 2006;
Ansell et al., 2007).
Speciation by hybridization is a popular issue among botanists exploring plant diversity; however, an important feature of speciation by hybridization is the formation of reproductive barriers between hybrids and parental species (cf. Rieseberg et al., 1995; Petit et al., 2004). An alternative consequence of hybridization is a wide range in genetic variation, resulting in the formation of a hybrid swarm (e.g. Wiens et al., 2006; Wachowiak and Prus-Glowacki, 2008). This may offer the potential for widening of ecological niches (Choler et al., 2004). In other words, successful hybridization may lead to either rapid speciation or elevated genetic variation. Several studies have shown that novel alleles might evolve in the context of hybridization (Schilthuizen et al., 1999; Schilthuizen et al., 2001; Smith et al., 2003). These novel alleles were widespread in the hybridized population
6These three authors contributed equally to this work.
*Corresponding authors: E-mail: yuchung@mail.nsysu.edu.tw, Tel: +886-7-5253625, Fax: +886-7-5253609 (Yu-Chung Chiang); E-mail: ypcheng@tfri.gov.tw, Tel: +886-2-23039978 ext. 2700, Fax: +886-2-23142234 (Yu-Pin Cheng).