Botanical Studies (2010) 51: 421-430.

Cloning, identification and characterization of a
repetitive sequence flanking telomere and homologous
to canrep in Brassica napus
repetitive sequence flanking telomere and homologous
to canrep in Brassica napus
Chen LI1,2, Bin HUANG3, Xiao-Hong YAN2, Li-Jun WANG2, Qing YANG1'*, and Wen-Hui WEI2 *
1College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China
2Institute of Oil Crops, Chinese Academy of Agricultural Sciences/Key Laboratory of Oil Crop Biology of the Ministry of
Agriculture, Wuhan 430062, China
3College of Life Sciences, Xinyang Normal University, Xinyang 464000, China
2Institute of Oil Crops, Chinese Academy of Agricultural Sciences/Key Laboratory of Oil Crop Biology of the Ministry of
Agriculture, Wuhan 430062, China
3College of Life Sciences, Xinyang Normal University, Xinyang 464000, China
(Received October 22, 2009; Accepted February 4, 2010)
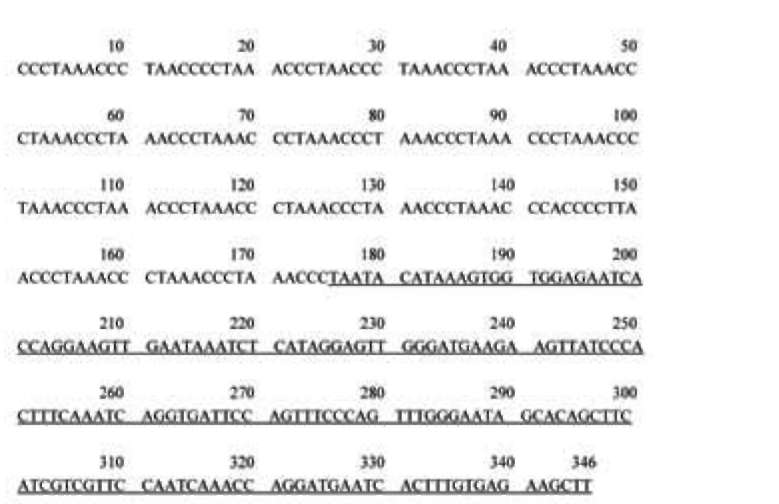
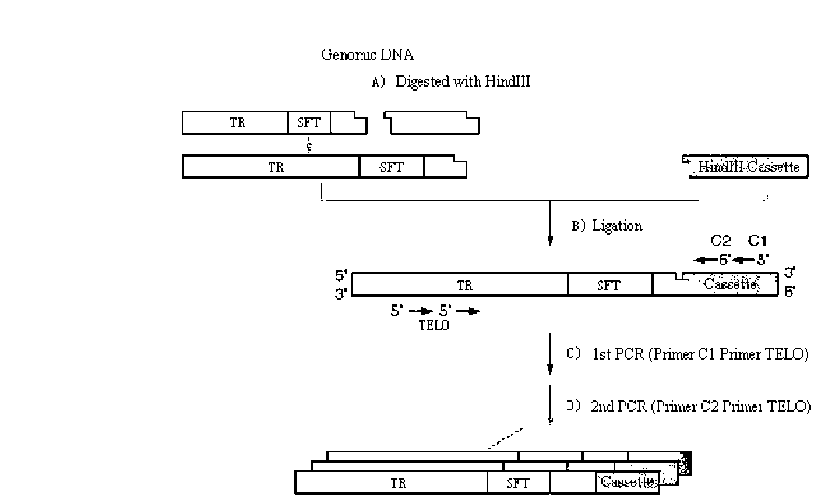
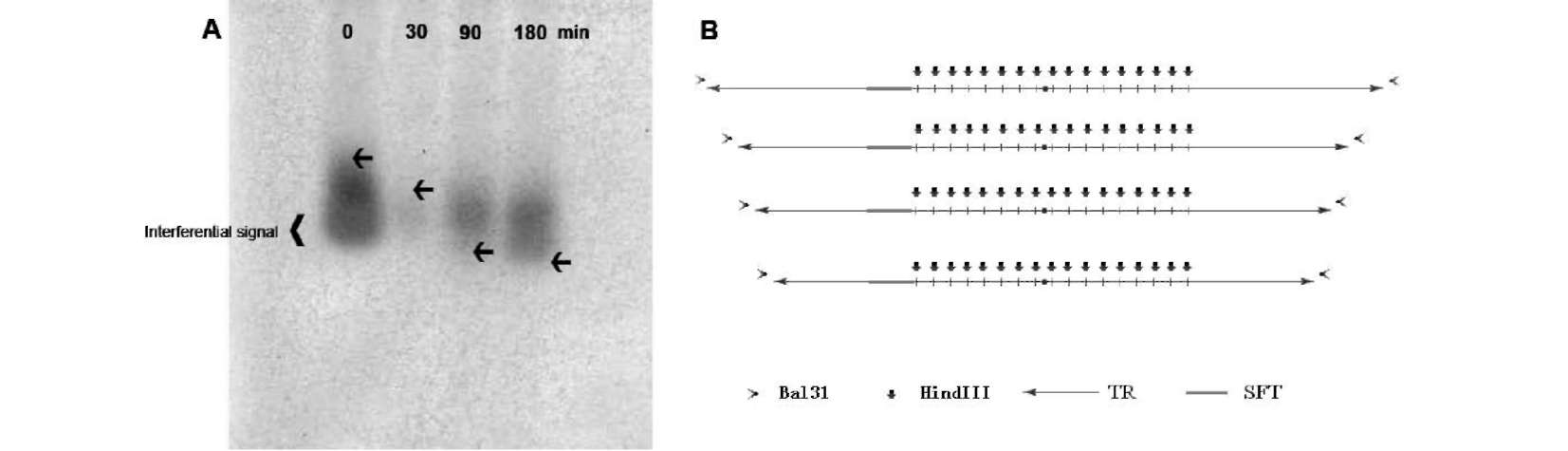
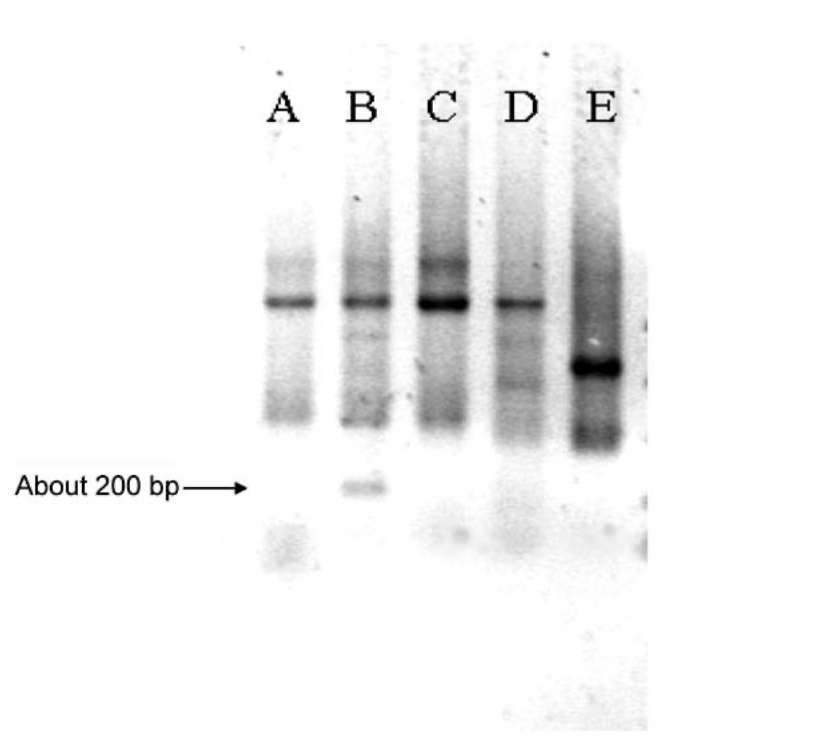
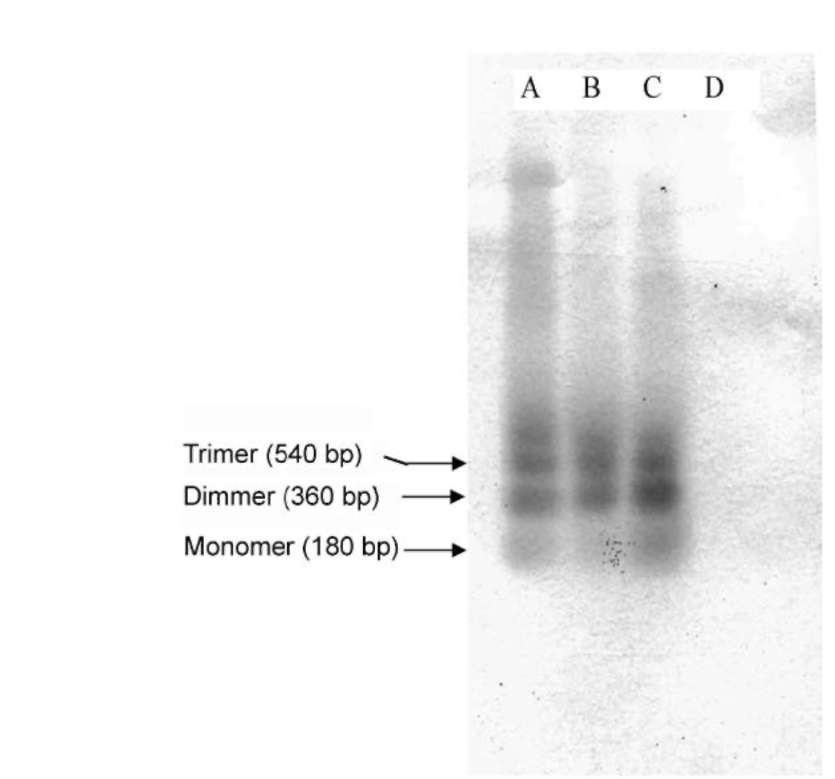
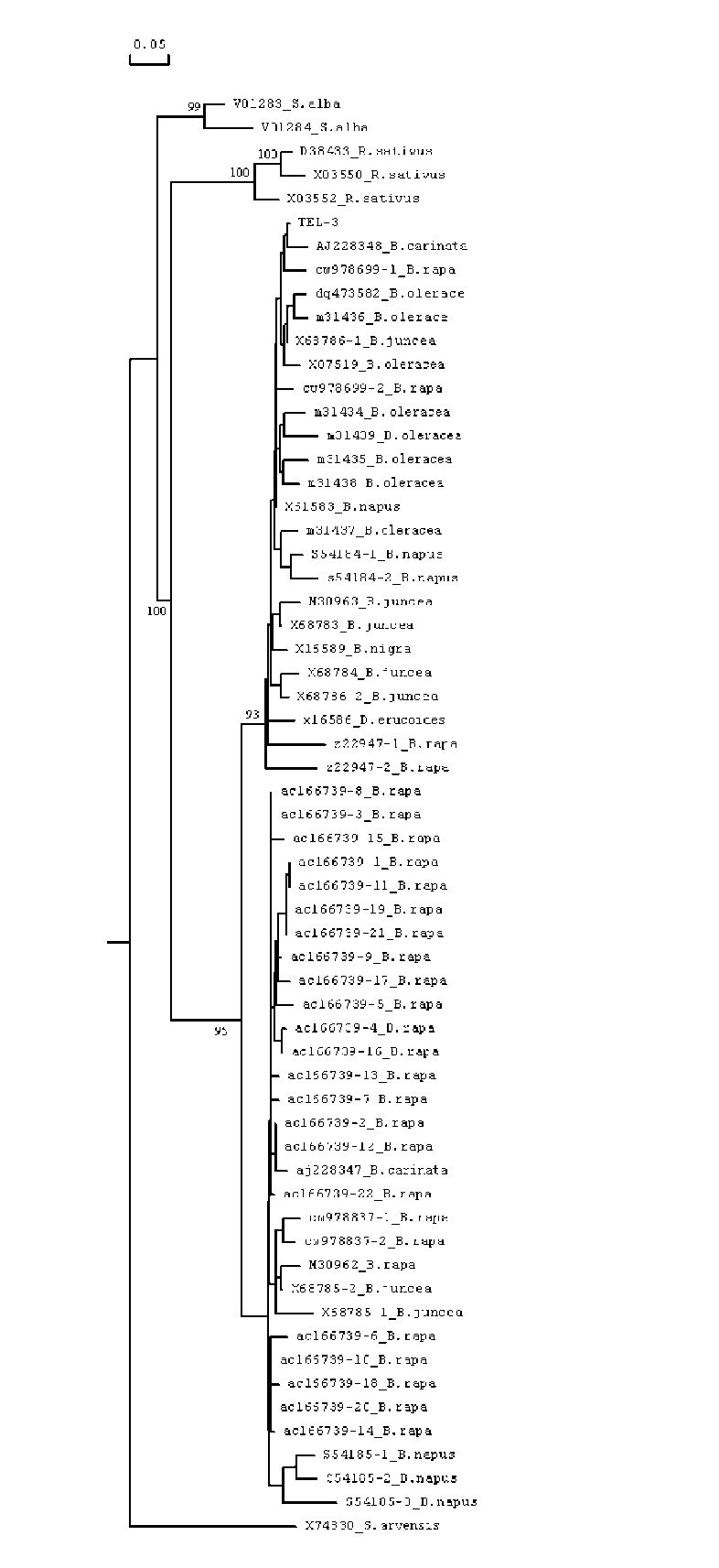
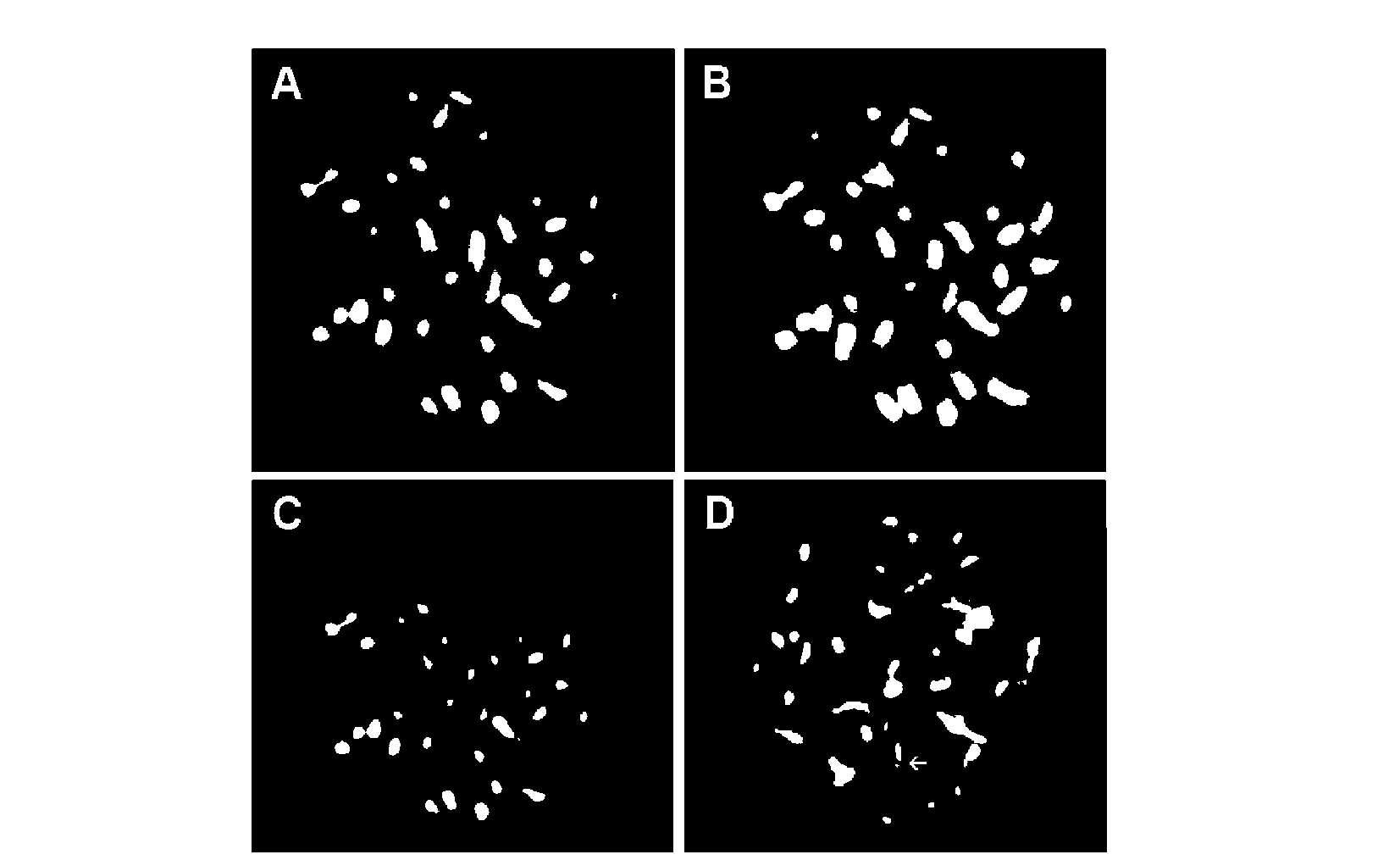
ABSTRACT. Satellite DNAs consist of long tandem arrays of short or large repeated sequences that form the centromeric and sometimes also subtelomeric regions of all higher eukaryote chromosomes. Telomeres and centromeres are the most important elements involved in the structure and function of chromosome. Telomeric DNA sequences are conserved among nearly all plant species. Although centromere-specific DNA sequences have been isolated in a wide range of plant species, almost no conservation was found in their DNA sequences. Here, we utilized the cassette-ligation-mediated PCR approach for cloning the sequence near the telomeric repeats of Brassica napus. Sequences analysis showed that one sequence was highly homologous to the centromeric satellite DNA sequence reported. FISH revealed that this satellite was located primarily at centromeric regions of most chromosomes, and also at some chromosome ends. The sequence was a subtelomeric satellite DNA which possessed variability in the evolution of chromosomes of Brassica species. The phylogenetic tree analysis of the satellite showed its sequence was conservative in Brassica. We discussed why the centromeric satellites appeared in the subtelomeric region.
Keywords: Brassica napus; Centromere; Satellite DNA; Subtelomere; Telomeric repeat.
INTRODUCTION
on the 3 '-end of each strand of chromosomal DNA. The telomeric and centromeric DNA, RNA, and protein components have been analyzed in recent years. Although our understanding of their functions remains elusive, recent findings show that they have some similarities. For instance, telomeric-like sequences are present in centromeric regions in Arabidopsis thaliana (Richards et al., 1991), maize (Alfenito et al., 1993), and potato (Tek and Jiang, 2004). There is a common telomeric-like secondary structure in Drosophila centromeric DNA (Abad et al., 2000). The centromeric and subtelomeric regions of eukaryotic chromosomes consist of mosaics of repeats and retrotransposons structured in a remarkably similar way (Pryde et al., 1997; Nagaki et al., 2004). 一
The subtelomeric regions of most organisms are dynamic with frequent turnover and exchange of sequences. In general, their structures are conservative from yeast to humans (Pryde et al., 1997). In plant, there are large tracts of tandem repeats in subtelomeric regions, often with spacer sequences between them and TRs (Ganal et al., 1992). The spacer sequences are called telomere-associated sequences (TASs). Telomere-associated regions represent boundaries between the relatively homogeneous telomeres and the subtelomeres, which show much greater heterogeneity in chromatin structure
The majority of genomic DNA in most plant species is made up of repetitive elements including satellites and retrotransposons. Satellite DNAs consist of long tandem arrays of short or large repeated sequences that form the centromeric regions of all higher eukaryote chromosomes. They are sometimes also found in subtelomeric or other chromosomal locations. Satellite DNAs are implicated in centromeric functions, such as segregation in mitosis and meiosis, recognition and pairing of homologous chromosomes, sister chromatid attachment, and formation of kinetochore structures (Willard, 1998). Telomeres and centromeres are the most important functional elements in plant chromosomes, as in other eukaryotic chromosomes. The two elements are in general composed of repetitive DNA sequences and binding or associated proteins (Murata, 2002). In most eukaryotic chromosomes, telomeres are composed of variable numbers of simple repeat sequences characterized by clusters of G residues
*Corresponding authors: E-mail: whwei@oilcrops.cn, Tel: +86-27-86722567, Fax :+86-27-86817881 (Wen-Hui WEI); qyang19@njau.edu.cn, telephone/Fax: +86-25-84395221 (Qing YANG).