UDDIN and KIM ― Plant RNAi spread
131
reporter fusions, Schwab et al. (2009) reported that AUXIN RESPONSE FACTOR (ARF) controlling ta-siRNAs behave non-cell autonomously to control ARF activities. Slotkin et al. (2009) proposed that siRNAs processed from trans-posable elements could also move from pollen vegetative nuclei to sperm cells to mediate transposon silencing in the germ line. Another intriguing result was recently published by Olmedo-Monfil et al. (2010) on the movement of 24 nt-long ta-siRNAs during female gametogenesis in Arabidop-sis. Thus, various small RNAs have emerged as potential mediators of non-cell autonomous RNA silencing.
SILENCING SIGNAL SPREAD BETWEEN
CELLS
The expansion of RNA silencing into neighboring plant cells can be divided into two distinct types of mechanisms, namely cell-to-cell and long-distance spread. Cell-to-cell spread consists of two sequential events, local (limited) and extensive silencing (Figure 1).
Local silencing spread
Local cell-to-cell transport of RNA silencing can be distinguished from extensive silencing spread by the activity of cellular RNA-dependent RNA polymerases (RDRs). In
the absence of amplification triggered by RDR activity, the extent of silencing through plasmodesmata is limited to as few as 10-15 cells beyond the site of initiation, which is known as 'local' or 'limited' cell-to-cell silencing spread. Plasmodesmata are unique intercellular channels found in most plant cells, with the exception of mature guard cells and cells at the maternal/filial boundary (Lucas and Lee, 2004; Oparka, 2004; Kim and Zambryski, 2005; Maule, 2008). Interestingly, the local movement mechanism relies on the DCL4-dependent production of 21-nucleotide siRNA when endogenous genes are targeted in an AGO1-dependent manner (Himber et al., 2003; Parizotto et al., 2004). Cell-to-cell spread of RNA silencing in Arabidopsis occurs during embryogenesis through distinct plasmod-esmata apertures (Kobayashi and Zambryski, 2007). Furthermore, using size-specific GFP tracers, the extent of silencing signal was found to be similar to that of soluble proteins between 27 to 54 kDa (Kobayashi and Zambryski, 2007).
When a double stranded SULPHUR gene fragment was expressed using a phloem companion cell-specific promoter, the non-cell autonomous silencing of the endogenous SULPHUR gene was manifested by a yellow chlorosis (reduction of chlorophyll concentrations) phenotype observed in companion cells and in the 10-15 adjacent cells
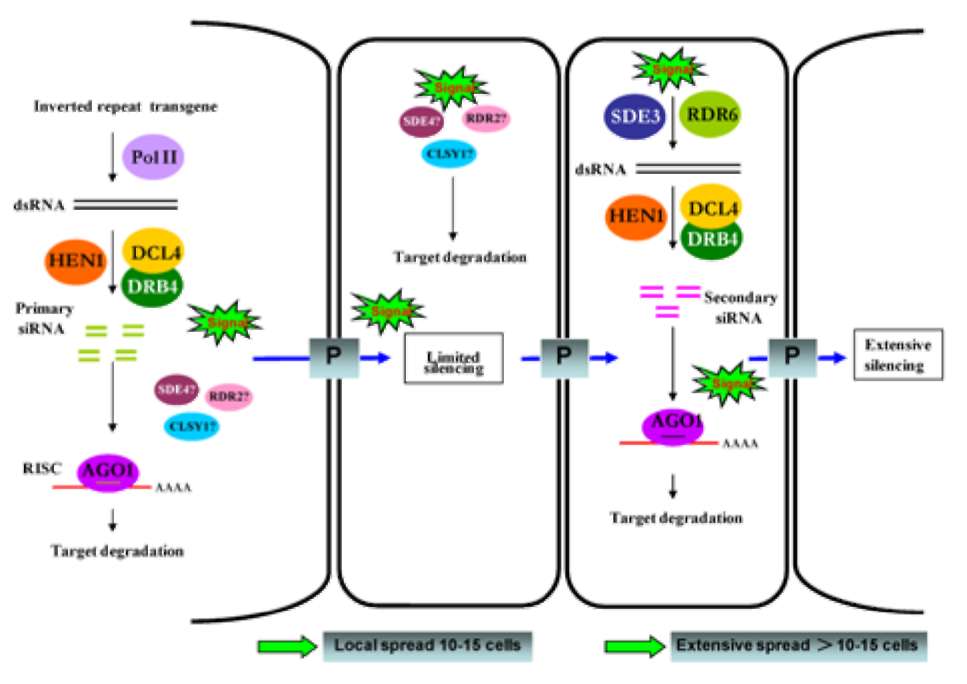
Figure 1. A simple model for inverted repeat transgene mediated non-cell-autonomous RNA silencing in plants. When an inverted repeat transgene is introduced into plants, dsRNAs are synthesized by polymerase II (Pol II), then cleaved by DCL4 with the cooperation of DRB4 to generate 21-nt long primary siRNAs. The siRNAs are stabilized by HEN1-mediated methylation and uploaded into effector protein complexes (RISCs), which degrade target mRNA in an AGO1-dependent manner. The 21nt siRNA, dsRNA or aberrant mRNA may able to move into neighboring (10-15) cells to enhance silencing machinery through plasmodesmata (P) by an unknown mechanism. SDE4/NRPD1a, RDR2, and CLSY1 have been reported to be involved in modifying the signal in donor cells or recipient cells via this local silencing pathway (details in text). Alternatively, an unknown signal could be amplified through the combined action of cellular RDR6 and SDE3 to synthesize new dsRNA, which is processed by the silencing machinery. This process was reiterated to induce extensive silencing spreads (more than 10-15 cells) in Arabidopsis through plasmodesmata. This model is based on Himber et al. (2003) and Dunoyer et al. (2007).