SHIEH et al. — OEC in marine diatom
163
PAGE. The polypeptidic bands were visualized either by coomassie blue or silver staining. The relative amount of the protein component on SDS-PAGE was determined by a densitometric analysis (Lee et al., 2006).
Purification of 33 kDa and 15 kDa proteins from diatom thylakoids
The 33 kDa protein was extracted from diatom thy-lakoids with 1.2 M Tris-HCl (pH 8.3) buffer after 0.8 M NaCl treatment, and the 15 kDa protein was extracted with 0.8 M NaCl. The crude extracts were concentrated by ultrafiltration. The concentrations of 33 kDa and 15 kDa proteins were determined by a modified Lowry method NaCl treatment, and the 15 kDa protein was extracted with 0.8 M NaCl. The crude extracts were concentrated by ultrafiltration. The concentrations of 33 kDa and 15 kDa proteins were determined by a modified Lowry method (Larson et al., 1980).
Measurement of Ca and Mn content in diatom thylakoids
The contents of Ca and Mn in diatom thylakoids were determined with an Inductively Coupled Plasma Emission Spectrometer (ICPAES). To remove salts, the Tris- or Na-Cl-washed diatom thylakoids were dialyzed with distilled deionized water overnight. Thylakoids were collected at 15,000 x g for 30 min and then dissolved with pure HNO3 and H2SO4 (4:1) just prior to measurement.
Reconstitution of Tris- and NaCl-washed diatom thylakoids
The Tris- or NaCl-washed diatom thylakoids were incubated with 33 kDa and/or 15 kDa proteins with or without
addition of 40 mM MnCl2 in buffer II at 4°C stirring for 1 h under room light of 10-20 fE m-1 sec-1. The protein to Chl content ratio was 2:1 (fg proteins versus fg Chl) in the reaction solution. The reconstituted thylakoids were then collected by centrifugation at 15,000 xg for 30 min.
Amino acid composition analysis
To prepare the 33 kDa and 15 kDa proteins for amino acid analysis, polypeptide bands on SDS-PAGE were visualized with 4 M sodium acetate in the dark (Hig-gins and Dahmus, 1979). The 33 kDa and 15 kDa bands were cut, chopped into pieces, homogenized, and then immersed into distilled water and stirred overnight. The eluted proteins were precipitated with 80% (v/v) acetone and dissolved with 6 M HCl in new tubes at 110°C for 24 h in the presence of 1% (v/v) phenol. The hydrolytes were subjected to analysis by an LKB4150 Alpha Acid analyzer (Taipei Regional Analytical Instrumentation Center).
RESULTS
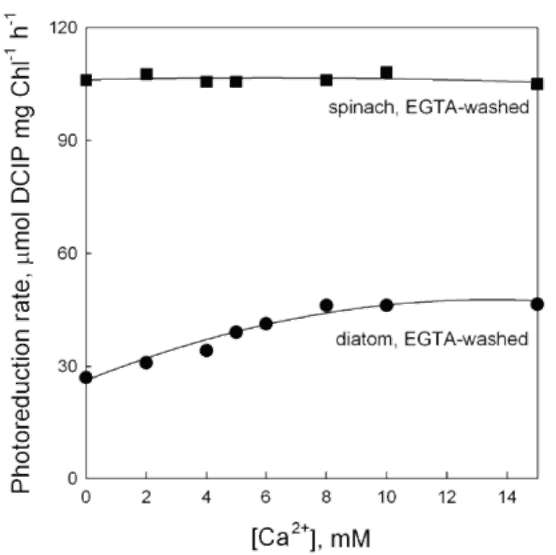
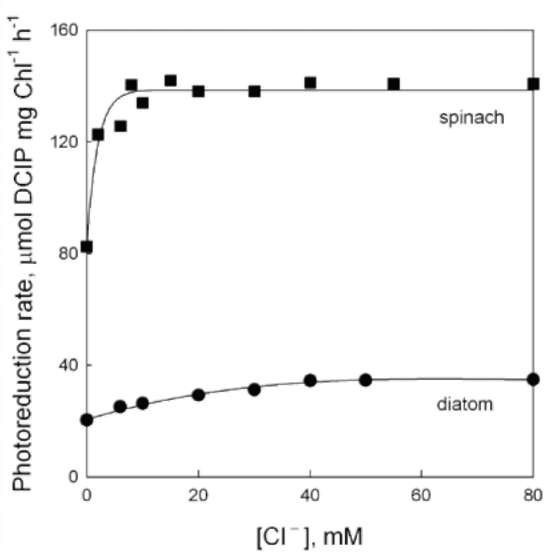
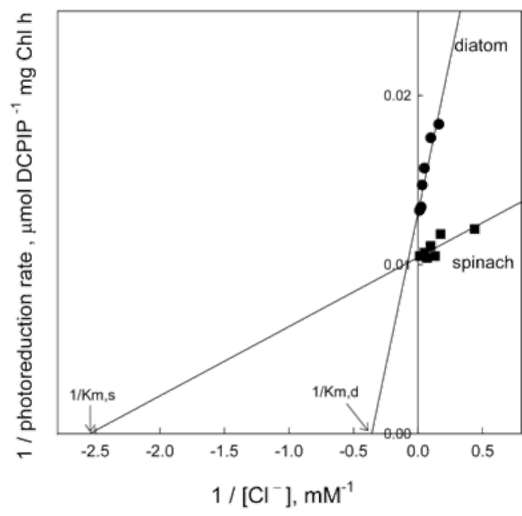
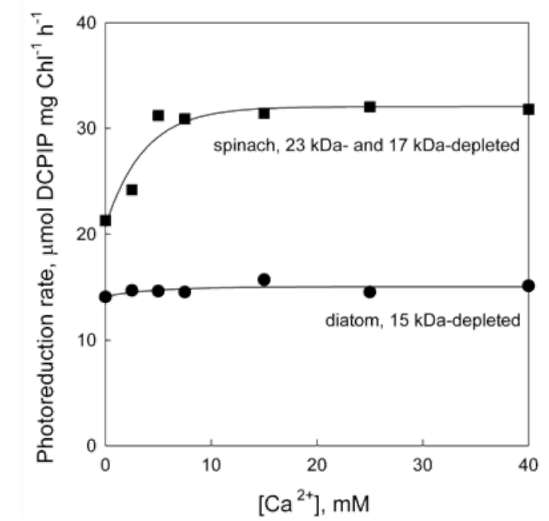
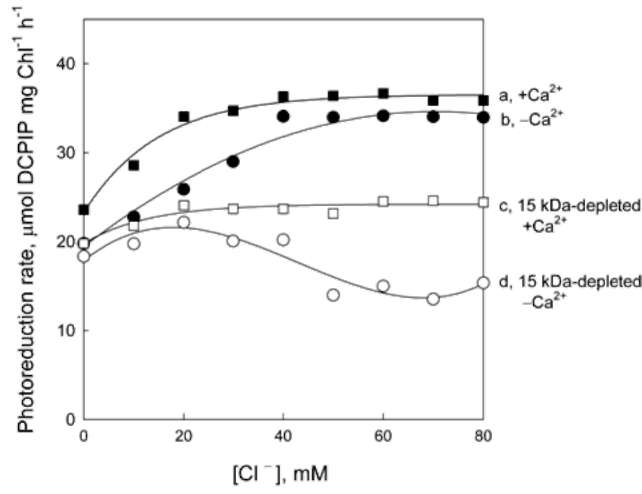
Effect of Tris and NaCl washing on electron transfer in the OEC of diatom thylakoids
The polypeptidic components in the OEC of diatom th-ylakoids were analyzed by SDS-PAGE and compared with those of spinach. In contrast to spinach thylakoids (Figure 1A, lane 1), diatom thylakoids showed two proteins of 33 kDa and15 kDa (molecular weights estimated in accordance with protein markers) but no 23 kDa protein (Figure
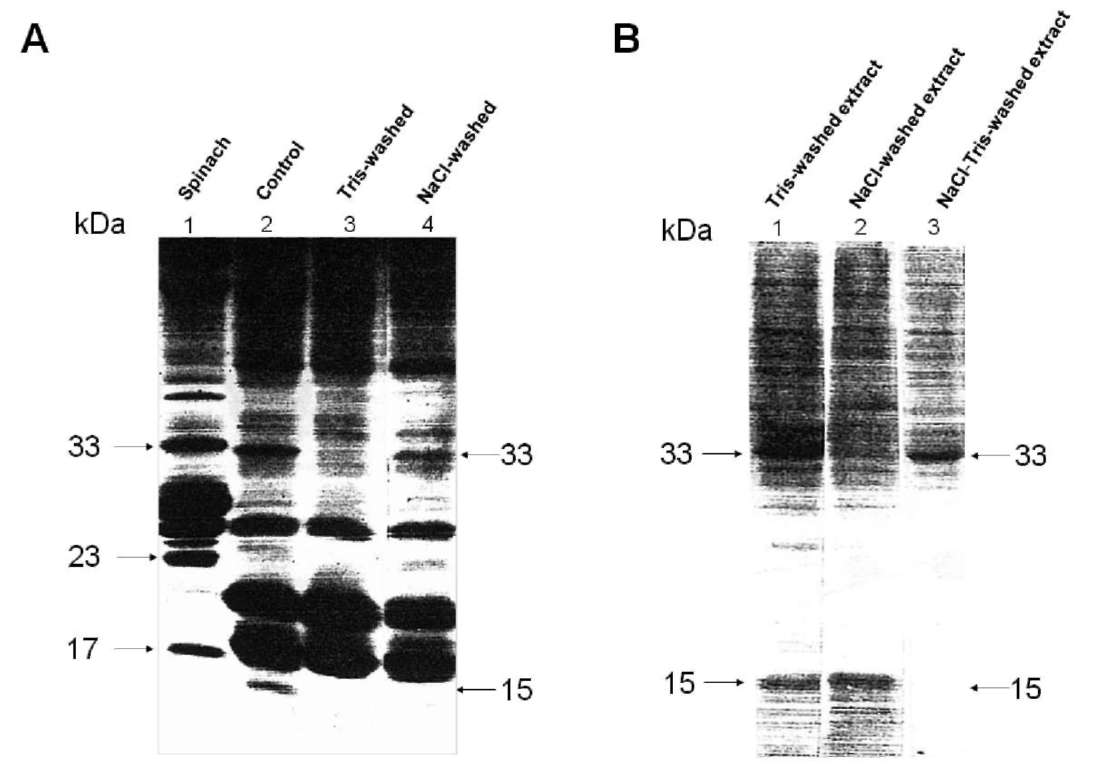
Figure 1. SDS-PAGE analysis of thylakoid membranes. (A) Lane 1, spinach control membranes; lane 2, diatom control membranes; lane 3, Tris-washed diatom membranes; lane 4, NaCl-washed diatom membranes. (B) Lane 1, Tris-washed diatom extract; lane 2, NaCl-washed diatom extract; lane 3, Tris-washed diatom extract after NaCl treatment.