234
Botanical Studies, Vol. 52, 2011
Table 3. Analysis of molecular variance (AMOVA) of Sphaeropteris brunoniana based on AFLP data.
|
Source of variation
|
d.f.
|
SSD
|
Variance components Absolute %
|
- p-statistics
|
P-value
|
|||
|
Total
|
|
|
|
|
|
|
||
|
Among populations
|
9
|
917.336
|
5.037
|
12.4
|
9st= 0.12
|
<0.01
|
||
|
Within populations
|
122
|
4355.982
|
35.705
|
87.6
|
|
|
||
|
Yunnan vs. Hainan-Laos
|
|
|
|
|
|
|
||
|
Among regions
|
1
|
433.508
|
5.641
|
13.0
|
9ct= 0.13
|
<0.05
|
||
|
Among populations within regions
|
8
|
483.828
|
1.894
|
4.4
|
9sc= 0.05
|
<0.01
|
||
|
Within populations
|
122
|
4355.982
|
35.705
|
82.6
|
9st= 0.17
|
<0.01
|
||
d.f.: degree of freedom; SSD: sum of squared deviations.
by AMOVA analysis (Table 3), which revealed that 87.6%
of the total genetic variance was attributed to intra-popula-tions and only 12.4% was partitioned among populations. Calculated from the value of Gst, the level of gene flow (Nm) among populations was 1.31. The genetic differentiation of this species was low between the regions of Yunnan and Hainan-Laos, with G尸0.06 and Nm= 3.92. Shannon's information index and AMOVA analysis showed that only 5.8% and 13.0% of the total variation occurred between the two regions, respectively. Although the divergences at population and regional levels were low, they were significant (^sf= 0.12, P< 0.01; cpct= 0.13, P< 0.05) (Table 3). The Mantel test revealed that the genetic differentiation of S. brunoniana in the investigated populations was directly related to physical distance, with r= 0.837 and P= 0.001.
Two major clusters of populations that correlated to their geographical distribution were identified in the UPG國 MA dendrogram (Figure 1). One cluster was composed of populations from Yunnan region, while the other contained populations from Hainan-Laos. In the Yunnan cluster, populations B and D collected from the west of Yunnan province were further clustered, while the others collected from the southeast of Yunnan were grouped. In the Hainan-Laos cluster, all the populations sampled from Hainan
province were clustered together. This result showed that the samples collected from areas that were geographically closer were grouped together and that the S. brunoniana distributed in the two regions were significantly different from each other.
DISCUSSION
High genetic diversity within the relict species
In this study, the genetic variation based on AFLP data was investigated for S. brunoniana. Despite its endangered status, unexpectedly high levels of genetic diversity were detected, with Ht= 0.333, H= 0.499 and Hs= 0.281, Hpop= 0.419. These values were much higher than those for the other two Cyatheaceae species in China: S. lepifera (Hs= 0.057 and Ht= 0.064, Chen, 1995) and A. spinulosa (Hs= 0.141, Cheng et al., 2008) from Taiwan using allozyme markers, A. spinulosa from mainland China using RAPDs (Hs= 0.0132, Hp。p=0.0136 and H尸0.0590, Hsp= 0.0560, Wang et al., 2004). The genetic parameters of S. brunoni-ana were also higher than the values reported for three other tree ferns from Costa Rica, based on allozyme data (Soltis et al., 1991). The different levels of genetic diversity for tree ferns may be explained, in part by the use of different molecular markers, and in part by the different sampling strategies, such as using the maximum geographic distance between sampled populations.
The genetic diversity within populations of S. brunoni-ana (Hs= 0.281) was also higher than the average value for plants (Hs= 0.23) compiled by Nybom (2004) based on AFLP literature, whereas it was close to the average values for long-lived (Hs= 0.25) and outcrossing species (Hs=0.27) based on RAPD data statistics (Nybom, 2004). S.brunoniana must thus be a long-lived species. Moreover, Soltis et al. (1991) reported the outcrossing trait in three tree ferns, viz. A.firma, Cyathea stipularis and Lophosoria quadripinnata. Except A. loheri, the six other Cyatheaceae species native to Taiwan have been examined and have tended toward intergametophytic mating, especially inter-gametophytic crossing (Chen, 1995; Chiou et al., 2000, 2003). Although there is no information on the reproductive biology of S. brunoniana in the literature, the results
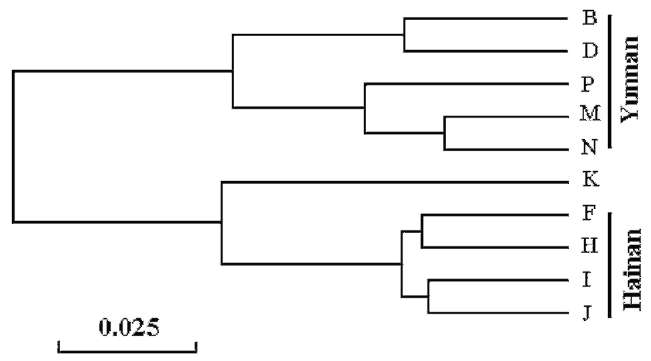
Figure 1. UPGMA dendrogram based on Nei's (1978) genetic distance showing relationships among Sphaeropteris brunoniana populations. Populations B, D, P, M and N located in Yunnan province. Populations F, H, I and J located in Hainan province and population K located in Laos.