WANG et al. ― Physiology of microalgal oil body formation
307
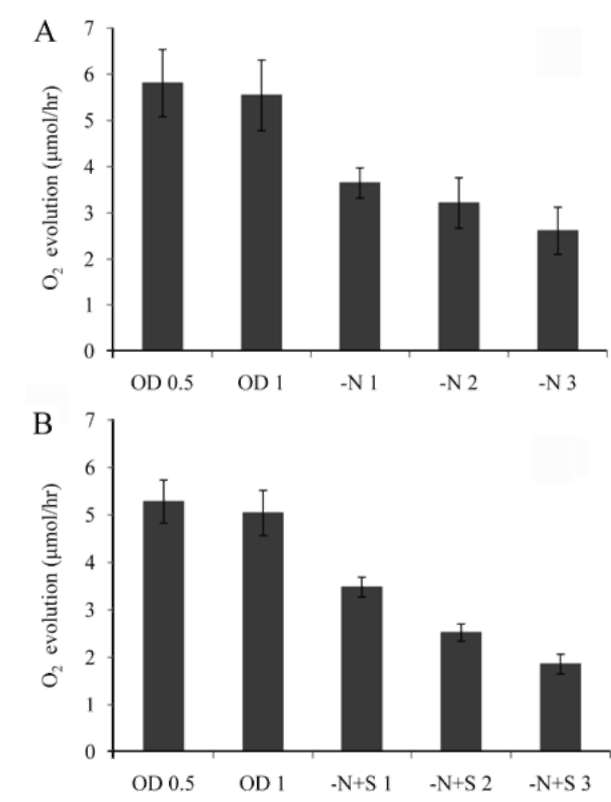
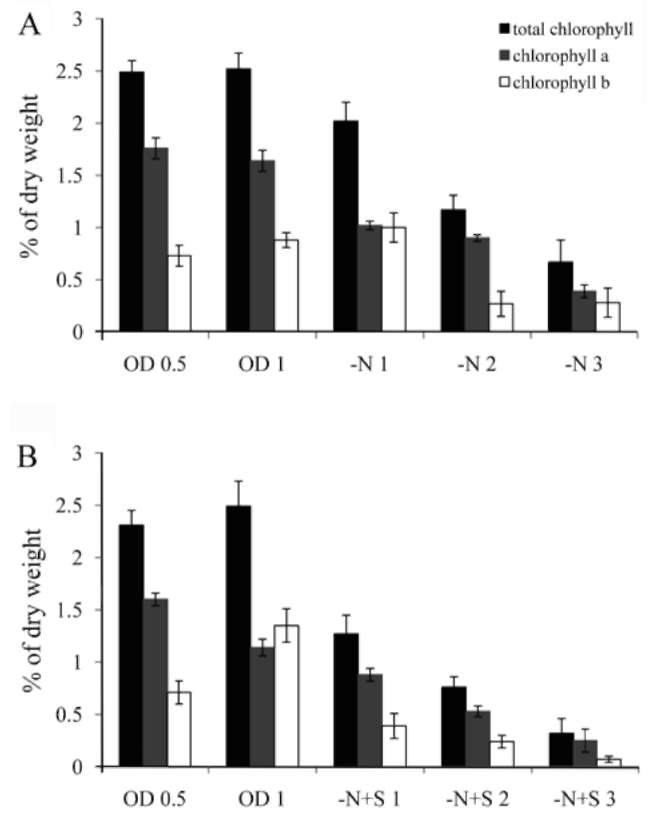
which was measured by a Clark-type oxygen electrode (Hansatech Inc. England). NaHCO3 solution was then added to the sample to reach 3 mM, and the oxygen evolution rate of these cells was measured for 10 min under 100 fmol/m2s light intensity at room temperature. DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea) was purchased from the Sigma company, USA (Cat. # D-2425). Chlorophyll of the cells was extracted using 80% acetone. After mixing by vortex for 20 min, the sample was stored at 4°C overnight. The debris was pelleted by centrifuga-tion, and absorbance at 645 and 663 nm of the supernatant was measured by a spectrophotometer. Chlorophyll content was calculated by the equation: Total chlorophyll (μg/mL) = 20.2(A645)+ 8.02(A663);Chlorophyll a (μg/ mL) = 12.7(A663)- 2.69(A645);Chlorophyll b (μg/mL)=
22.9(A645)- 4.68(A663).
tively), and the ITS sequences were used to BLAST the GenBank. The results showed that the most closely related species to this green alga are members of the Scenedesmaceae family rather than the Chlorellaceae family, which comprises Chlorella. The ITS2 sequences of 12 closely related species representing 12 genera were aligned, and a phylogenetic tree was constructed based on the alignment using the Neighbor-Joining method. As shown in Figure 1, UTEX 2219-4 is most closely related to Neodesmus danubialis, which is a species in the Scenedesmaceae. Of the 12 species, two are mosses (Plagiomnuim and Imbribryum), one is a member of the Haematococcaceae family (Haematococcus) and the remaining are within Scenedesmaceae.
It is not surprising to find that mosses are closely related to green algae, it has already been well accepted that mosses are descendents of a green algal species. On the other hand, one might wonder how the UTEX 2219-4, that is supposed to be a Chlorella species, can be closely related to members belonging to Scenedesmaceae and Haema-tococcaceae, rather than to Chlorellaceae. The taxonomy of green algae has been found increasingly inaccurate as more microalgal genomes are sequenced. For example, Chlorella vulgaris C-169 has been renamed Coccomyxa sp. C-169 after the Joint Genome Institute sequenced its genome (http://genome.jgi-psf.org/Chlvu1/Chlvu1.home. html). Therefore, there is a need to regroup microalgal taxonomy based on molecular markers.
Transmission Electron Microscopy (TEM)
The algal cells were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.0 at 4°C for 4 hours. After three 20 min buffer rinses, the samples were postfixed in 1% OsO^ in the same buffer for 4 hours at room temperature and then rinsed in three 20 min changes of buffer. Samples were dehydrated in an acetone series, embedded in Spurr's resin, and sectioned with a Lecia Reichert Ultracut S or Lecia EM UC6 ultramicrotome. The ultra-thin sections (70-90 nm) were stained with uranyl acetate and lead citrate. A Philips CM 100 transmission electron microscope at 80 KV was used for viewing.
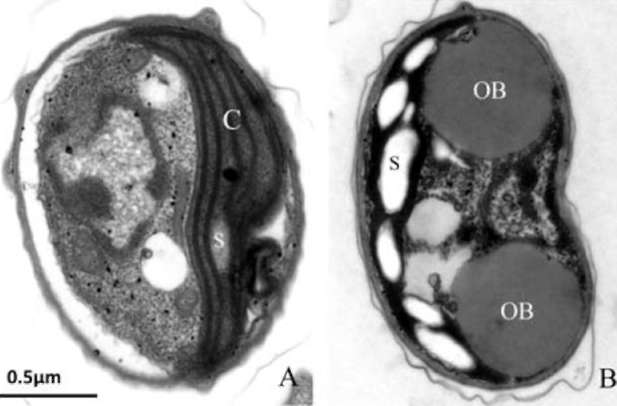
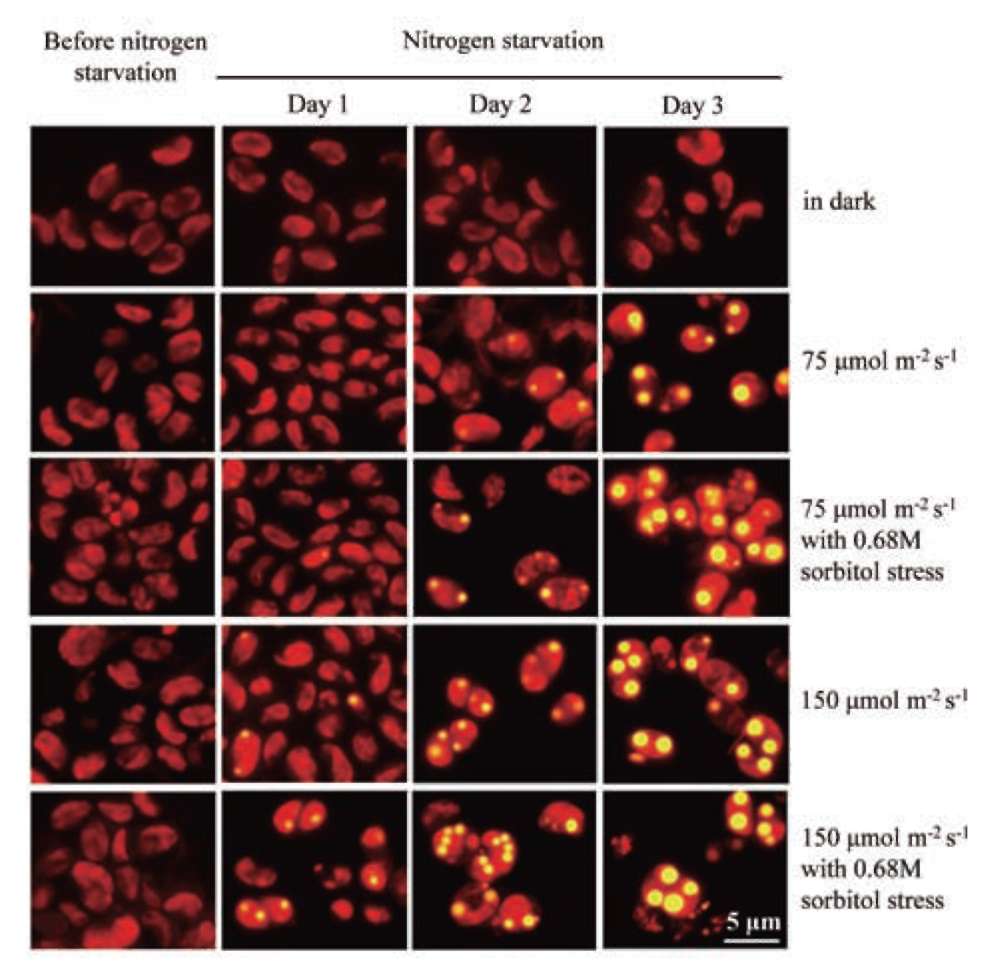
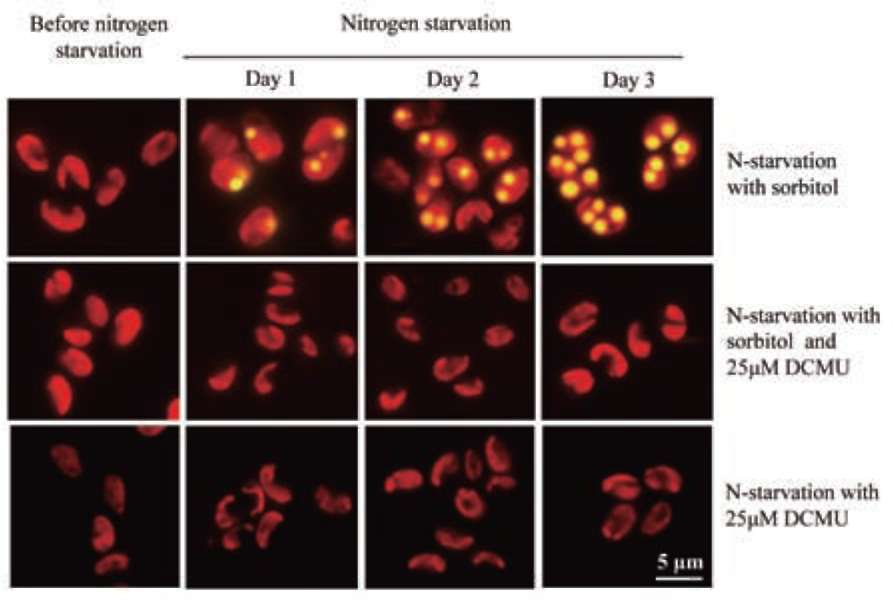
UTEX 2219-4 contained both starch granules and oil bodies after three days of nitrogen starvation
RESULTS AND DISCUSSION
UTEX 2219-4 is closely related to members in Scenedesmaceae
Most green microalgae produce starch as an energy reservoir under normal conditions. In nitrogen-deficient conditions, they start to accumulate triacylglycerol, which is stored in oil bodies (Moellering and Benning, 2010; Wang et al., 2009; Li et al., 2010). A few possible energy and carbon sources for the triacylglycerol biosynthesis at this
The ITS1-5.8S-ITS2 and the 18S rDNA regions of
UTEX 2219-4 were amplified and sequenced (GenBank accession numbers HQ218939 and HQ218940, respec-
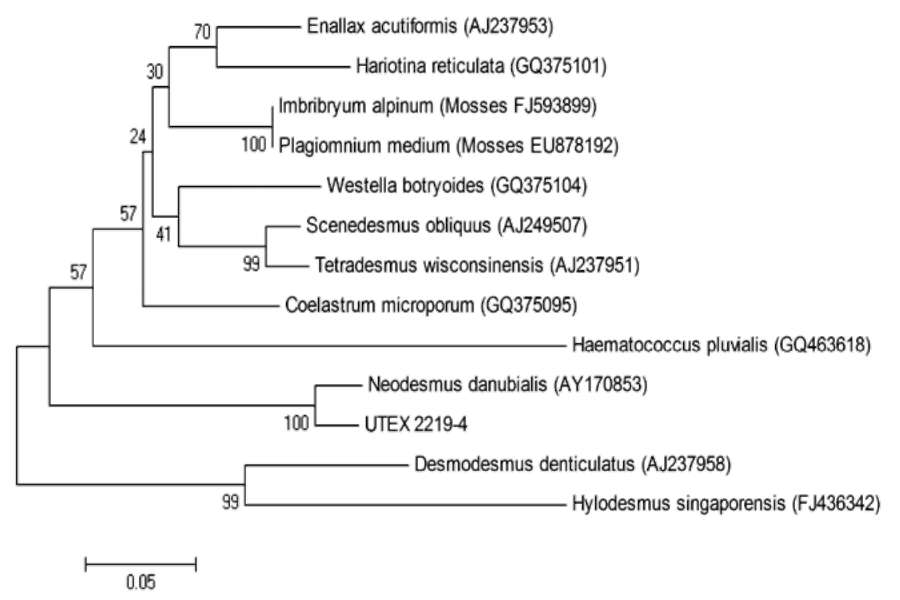
Figure 1. Phylogenetic analysis of UTEX 2219-4 and its closely related species based on ITS2 sequences in GenBank. ITS1 and ITS2 sequences of UTEX 2219-4 were used to BLAST in GenBank, and the 12 most closely related species, representing 12 genera, were selected for phylogenetic analysis. This tree was constructed based on the well-aligned regions in ITS2 sequences of each species. This tree showed that UTEX 2219-4 is closely related to species in the family Scenedesmaceae rather than to Chlorellaceae species. The numbers in the parenthesis are the accession numbers of the respective ITS sequences in GenBank. The scale bar measures the distance between species. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches.