LIU et al. ― Lily anther-specific genes
385
RESULTS
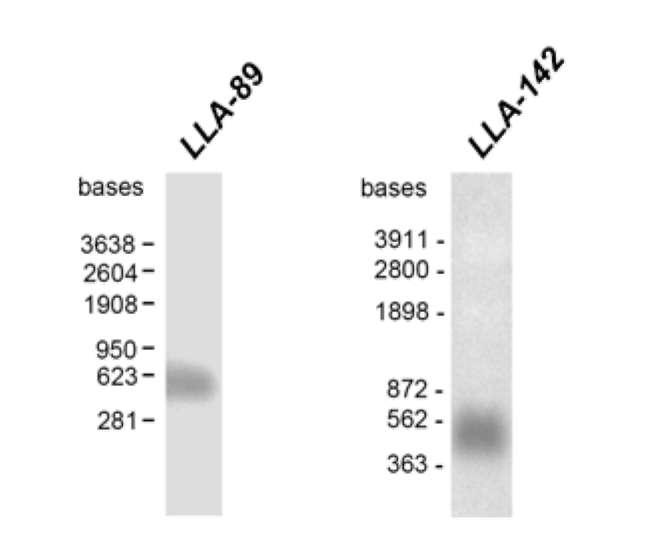
RACE-PCR method was used to obtain the full length 493 bp LLA-89 cDNA (accession no. DQ907930) and the full length 379 bp LLA-142 cDNA (accession no. EF026007), both excluding the poly (A) tail.
The LLA-89 cDNA contains an open reading frame of 303 bp encoding a polypeptide of 100 amino acids with a calculated molecular mass of 10.2 kDa and a pI of 3.2 (Figure 1A). The LLA-89 cDNA insert is close to full-
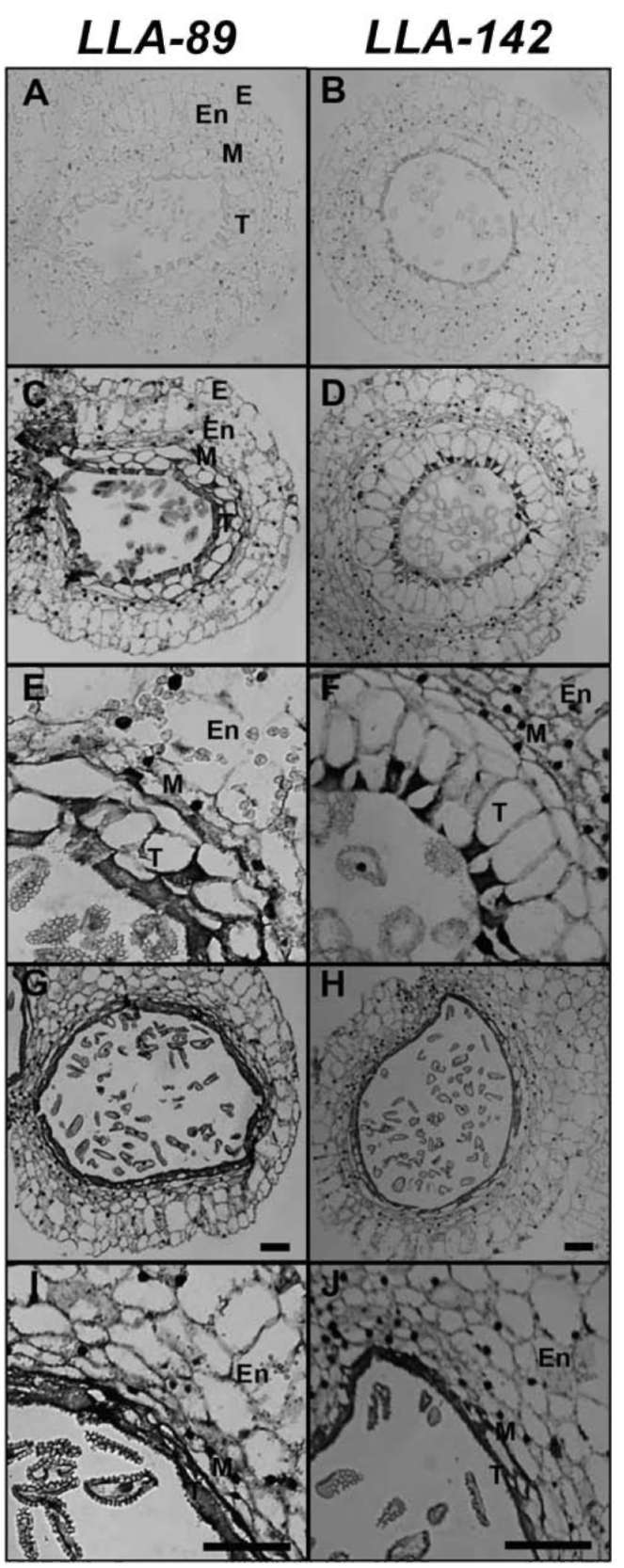
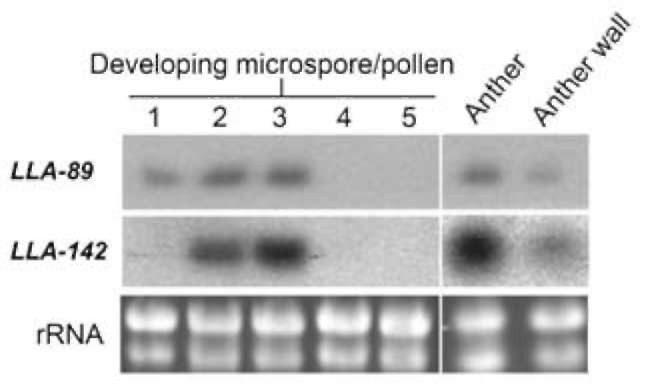
Characterization of the two stage-specific cDNAs in the anther of L. longiflorum
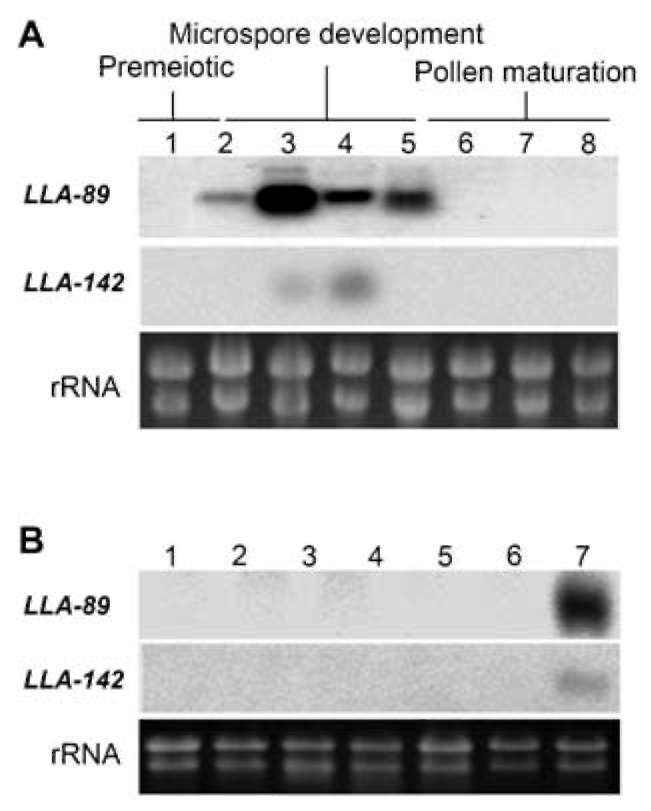
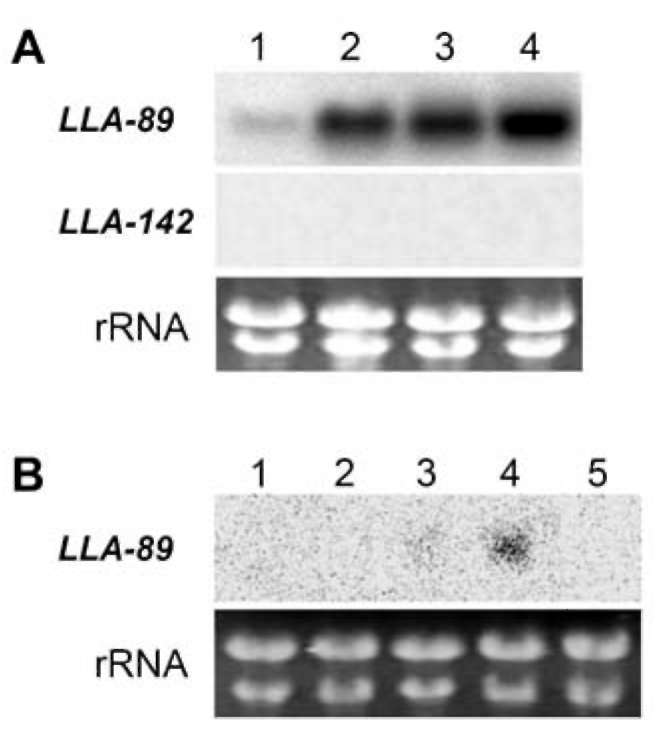
The LLA-89 and LLA-142 cDNAs were identified from a subtractive cDNA library at the L. longiflorum anther microspore development phase (Hsu et al., 2008). Since the insert sizes of both cDNAs were partial, the 5' - and 3'-
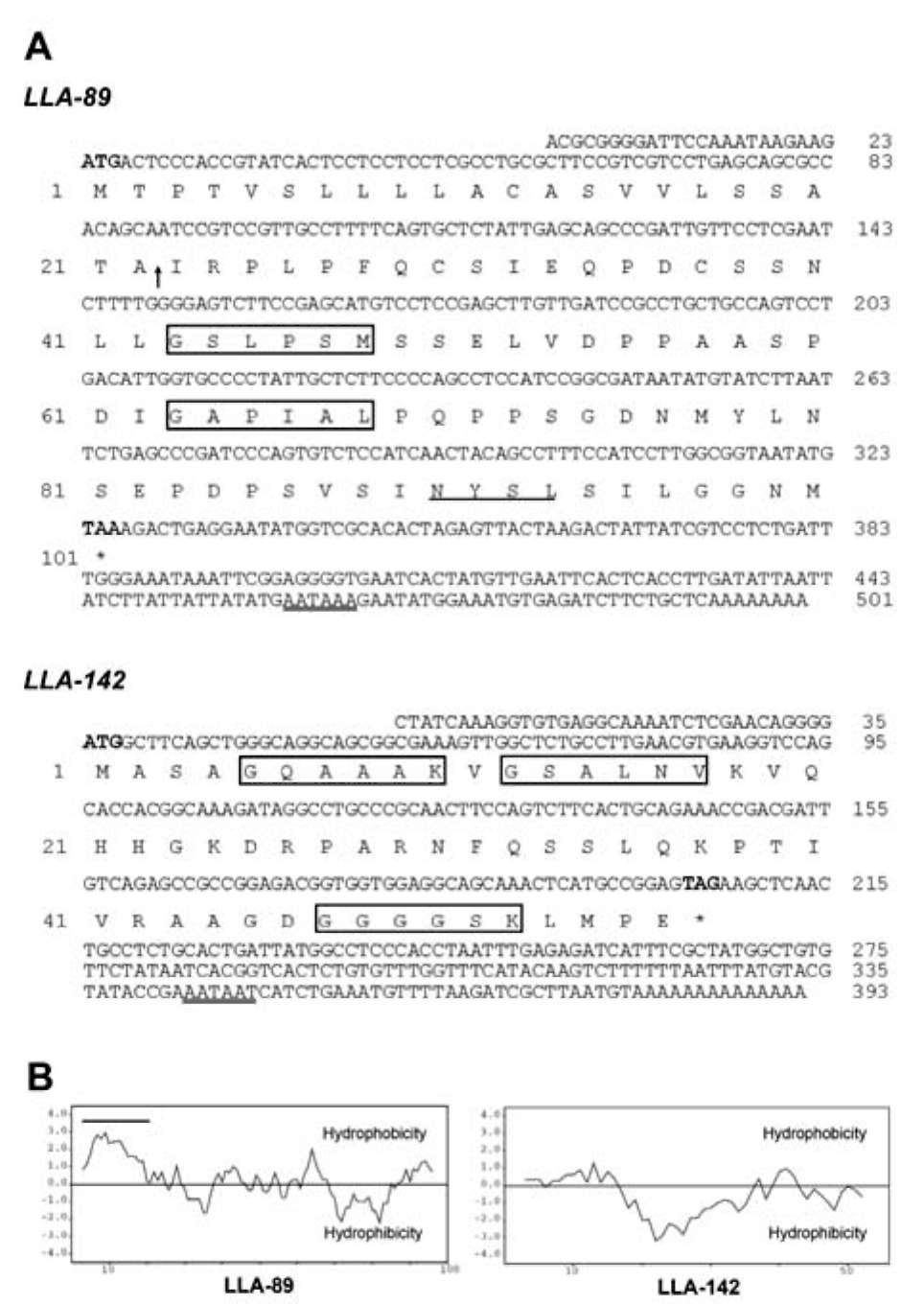
Figure 1. Sequences and hydropathy profiles of LLA-89 and LLA-142 in L. longiflorum anther. (A) Nucleotide and predicted amino acid sequences of LLA-89 and LLA-142 cDNAs. Numbers of nucleotide and amino acid sequences are indicated on the right and left, respectively. Bold letters in the nucleotide sequence indicate start codon and stop codon. The arrow indicates the predicted cleavage site of the signal peptide. The double underlines indicate the putative polyadenylation signal site. The underline indicates the putative N-glycosylation site (N-X-S/T). The putative N-myristoylation sites (G-X-X-X-S/T/A/G/C/N) are boxed; (B) Hydropathy profiles (Kyte and Doolittle, 1982) of LLA-89 and LLA-142 protein sequences. The black line indicates a hydrophobic sequence at the N-terminus of the sequence.