396
Botanical Studies, Vol. 52, 2011
ers, 1X TAKARA rTaq buffer and 0.25U TAKARA rTaq. PCR reactions were performed on a TAKARA PCR Ther-mal Cycler Dice Model TP600 (TaKaRa Bio Inc., Japan) for 1 cycle of 5 minutes at 94。C, followed by 35 cycles of 30 seconds at 94。C, 30s at annealing temperature, 30 sec-onds at 72°C, and a final 7-minute extension at 72°C. The optimal annealing temperature for all SSR primer sets was determined by gradient PCR condition conducted using a TAKARA PCR Thermal Cycler.
size variation of microsatellite fragments greater than 3 bp were considered as significantly different alleles.
Data analysis
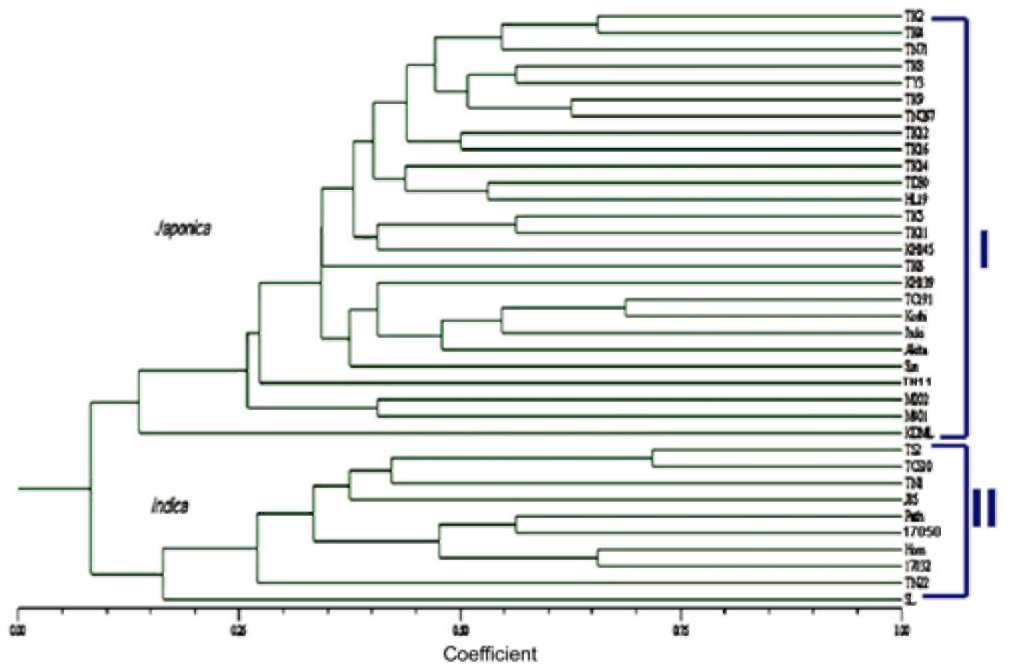
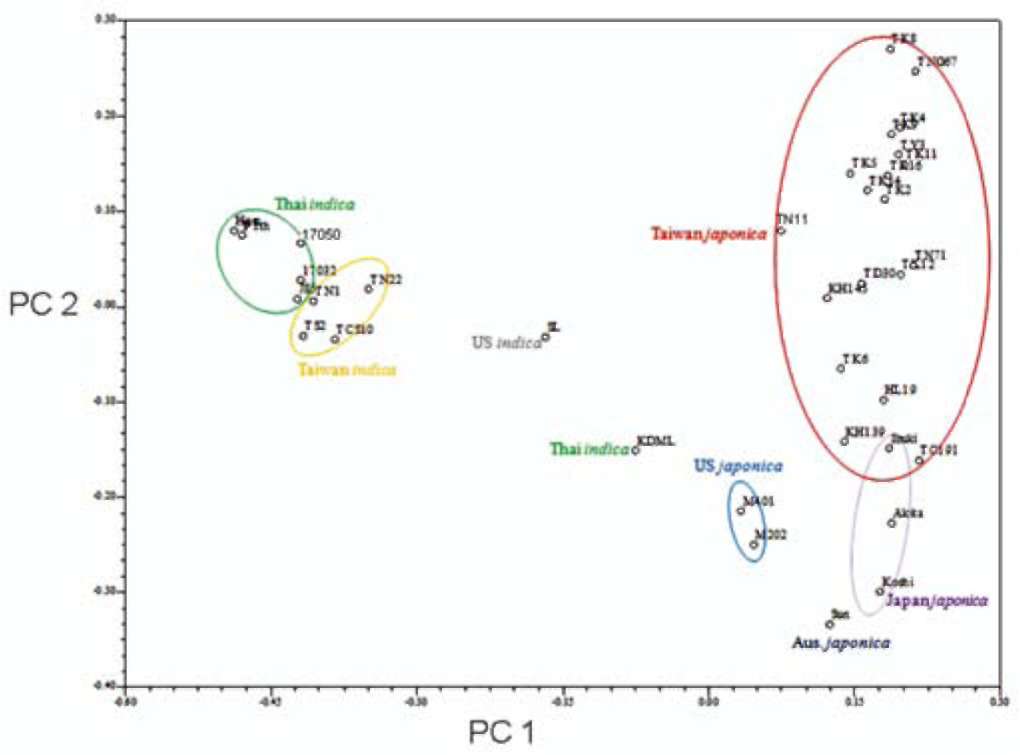
Molecular data from SSR assays, including genetic distance, cluster and principle component analysis (PCA), were analyzed with NTSYS pc ver 2. 2 (Exeter software). PCR products were scored as present (1) and absent (0) for each marker to generate a binary matrix. The genetic similarity of 23 local Taiwan rice cultivars was calculated, and a phylogenetic tree was constructed using the UPGMA (unweighted pair-group method using arithmetic averages) method based on genetic similarity coefficients estimated by Modified Rogers' Distance (RMD) (Goodman and Stu-ber, 1983; Reif et al., 2003; Reif et al., 2005):
PCR product separation and allelic size determination
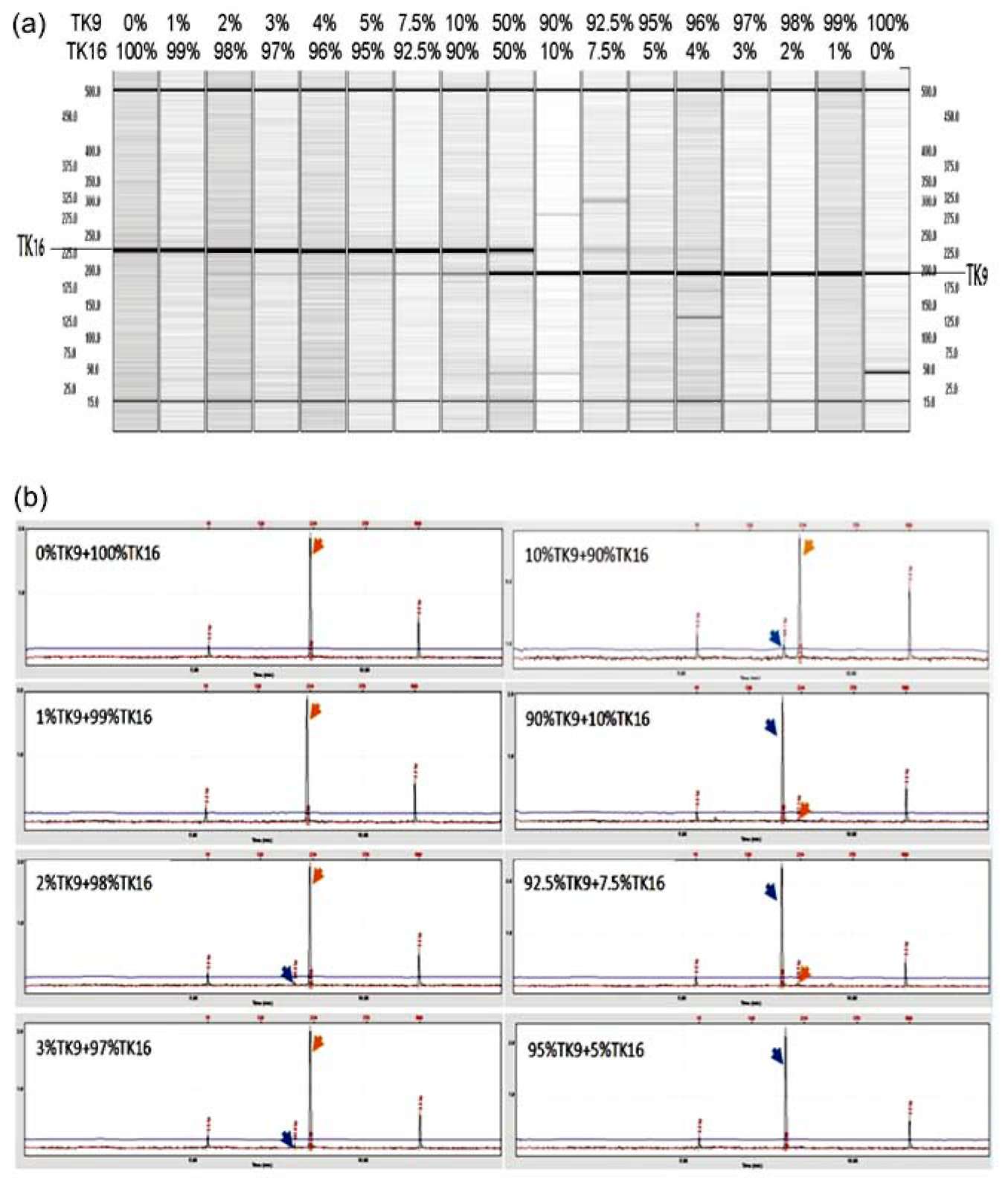
For optimal polymorphic SSR marker screening, PCR products were electrophoresised in 3% Metaphor agarose gel (Cambrex Bio science Rockland Inc., USA) with 1X TBE buffer, stained with EtBr and photographed under UV light. The SSR markers with high polymorphism were further used in SSR fingerprinting analysis and PCR products were analyzed on a QIAxcel multicapillary electrophoresis system (Qiagen; formerly eGene's HDA-GT12 system). Reactions were carried out with QIAxcel DNA high resolution Kit (Cat No. 929002) and the PCR products were separated using the OM750 method in a 12-channel, sieving-gel cartridge (GCK5000). Based on QX Align-ment Markers 15 bp/500 bp (Cat. No. 929520) and QX DNA Size Marker 25-450 bp (Cat No. 920555), actual al-lele sizes were resolved and automatically calculated in bp by BioCalculatorTM software, provided with the QIAxcel system, and shown as a gel image and electrophoregram. To fit into the best resolution capability of the system, the
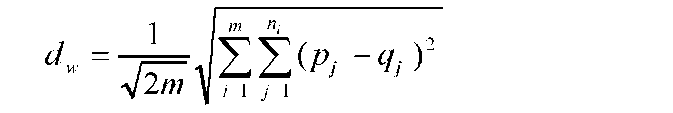
where and qy are the frequency of the jth allele at the ith marker in two independent cultivars and m, n indicate the number of markers. The polymorphic information content (PIC) for each marker was calculated (Botsein et al., 1980) and modified by Anderson et al. (1993) for self-pollinated species as follows:

where pi is the frequency of the ith allele, and n is the number of alleles.
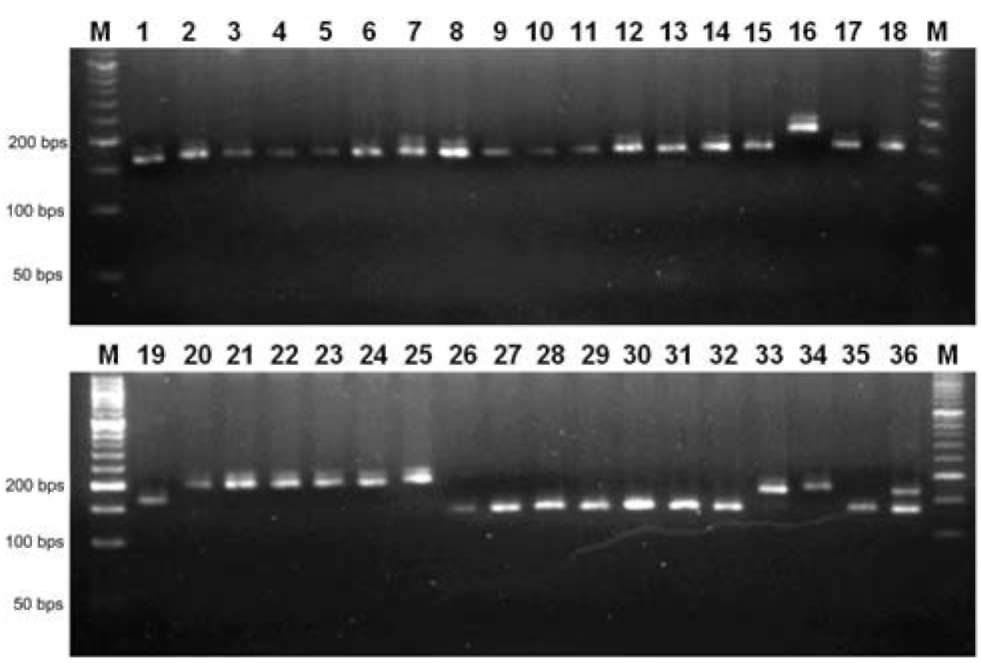
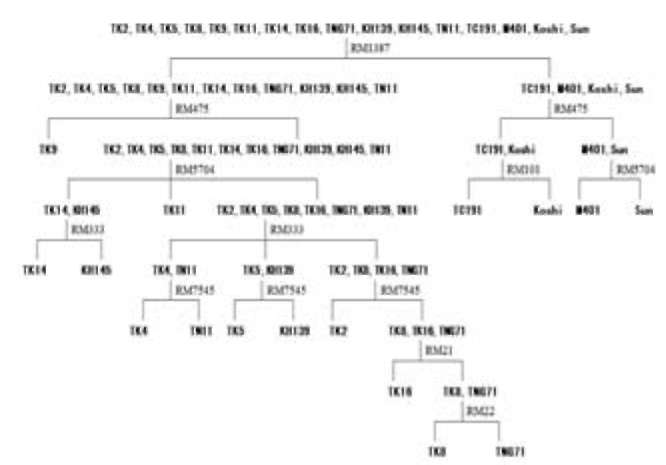
Figure 1. DNA fingerprint analysis of 36 rice varieties from different countries that were amplified by SSR marker (RM1387). The lane (M) represents 50 bp DNA molecule weight markers. The abbreviation name of rice cultivars used for this analysis is the same as Table 1. (1) TK2, (2) TK4, (3) TK5, (4) TK6, (5) TK8, (6) TK9, (7) TK11, (8) TK12, (9) TK14, (10) TK16, (11) TNG67, (12) TN71, (13) KH139, (14) KH145, (15) TN11, (16) TC191, (17) TY3, (18) TD30, (19) HL19, (20) M202, (21) M401, (22) Koshi, (23) Akita, (24) Ibuki, (25) Sun, (26) TS2, (27) TCS10, (28) TN22, (29) TN1, (30) J85, (31) Path, (32) Hom, (33) K17032, (34) K17050, (35) SL, (36) KDML.