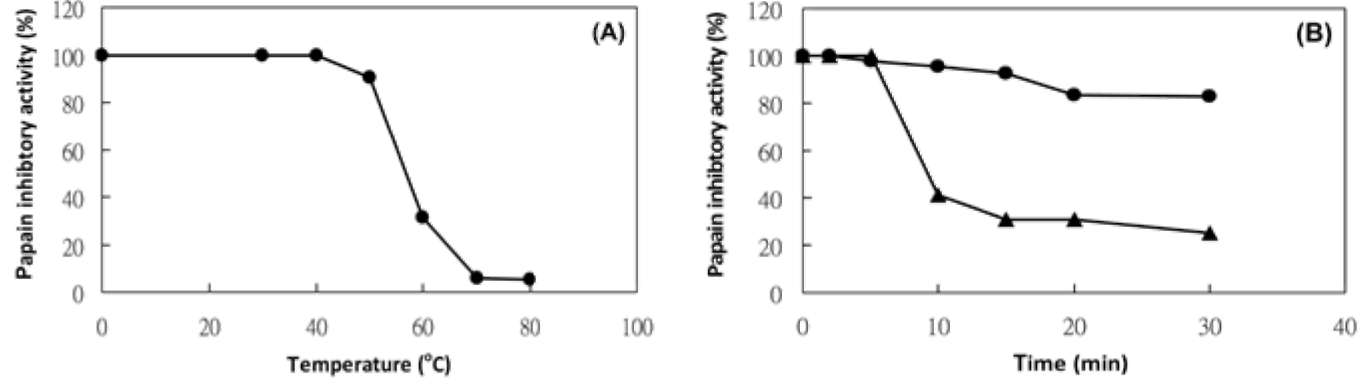
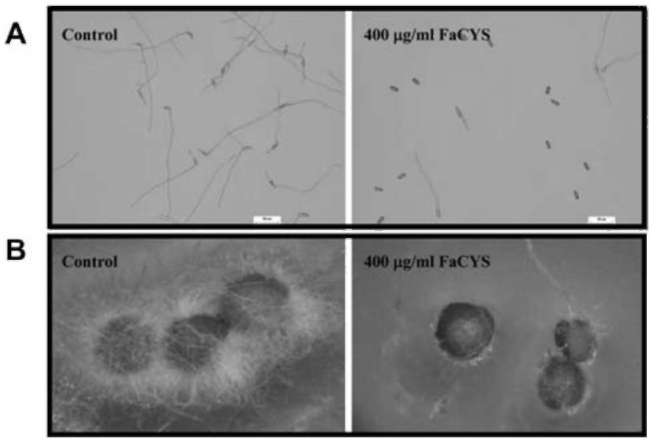
|
|
achenes. Biosci. Biotechnol. Biochem. 72: 506-513.
Corr-Menguy, F., F.J. Cejudo, C. Mazubert, J. Vidal, C. Le-landais-Biere, G. Torres, A. Rode, and C. Hartmann. 2002. Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Mol. Biol. 50: 687698.
Dixon, M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55: 170-171.
Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953-971.
Fabienne, C.M., J.C. Francisco, M. Christelle, V. Jean, L.B. Christine, T. Gisele, R. Andre, and H. Caroline. 2002. Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Mol. Biol. 50: 687-698.
Gaddour, K., J. Vicente-Carbajosa, P. Lara, I. Isabel-Lamoneda, I. Diaz, and P. Carbonero. 2001. A constitutively cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol. Biol. 45: 599-608.
Gianotti, A., W.M. Rios, A. Soares-Costa, V. Nogaroto, A.K. Carmona, M.L. Oliva, S.S. Andrade, and F. Henrique-Silva. 2006. Recombinant expression, purification, and functional analysis of two novel cystatins from sugarcane (Saccharum officinarum). Protein Exp. Purif. 47: 483-489.
Gianotti, A., C.A. Sommer, A.K. Carmona, and F. Henrique-Silva. 2008. Inhibitory effect of the sugarcane cystatin CaneCPI-4 on cathepsins B and L and human breast cancer cell invasion. Biol. Chem. 389: 447-453.
Girard, C., D. Rivard, A. Kiggundu, K. Kunert, S.C. Gleddie,
C. Cloutier, and D. Michaud. 2007. A multicomponent, elicitor-inducible cystatin complex in tomato, Solanum ly-copersicum. New Phytol. 173: 841-851.
Gutierrez-Campos, R., J.A. Torres-Acosta, L.J. Saucedo-Arias, and M.A. Gomez-Lim. 1999. The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in tran-sgenic tobacco plants. Nature Biotechnol. 17: 1223-1226.
Kondo, H., K. Abe, Y. Emori, and S. Arai. 1991. Gene organization of oryzacystatin-II, a new cystatin superfamily member of plant origin, is closely related to that of oryzacystatin-I but different from those of animal cystatins. FEBS Lett. 278: 87-90.
Kondo, H., K. Abe, I. Nishimura, H. Watanable, Y. Emori, and S. Arai. 1990. Two distinct cystatin species in rice seeds with different specificities against cysteine proteinase. J. Biol. Chem. 265: 15832-15837.
Martinez, M., I. Cambra, L. Carrillo, M. Diaz-Mendoza, and I. Diaz. 2009. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed
germination. Plant Physiol. 151: 1531-1545.
Martinez, M., M. Diaz-Mendoza, L. Carrillo, and I. Diaz. 2007. Carboxy terminal extended phytocystatins are bifunctional
inhibitors of papain and legumain cysteine proteinases.
FEBS Lett. 581: 2914-2918.
|
Nagata, K., N. Kudo, K. Abe, S. Arai, and M. Tanokura. 2000.
Three-dimensional structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 39: 14753-14760.
Neuteboom, L.W., K.O. Matsumoto, and D.A. Christopher. 2009. An extended AE-Rich N-terminal trunk in secreted pineapple cystatin enhances inhibition of fruit bromelain and is posttranslationally removed during ripening. Plant Physiol. 151: 515-527.
Nissen, M.S., G.N. Kumar, B. Youn, D.B. Knowles, K.S. Lam, W.J. Ballinger, N.R. Knowles, and C. Kang. 2009. Characterization of Solanum tuberosum multicystatin and its structural comparison with other cystatins. Plant Cell. 21: 861875.
Ohtsubo, S., M. Taiyoji, T. Kawase, M. Taniguchi, and E. Saitoh. 2007. Oryzacystatin-II, a cystatin from rice (Oryza sativa L. japonica), is a dimeric protein: possible involvement of the interconversion between dimer and monomer in the regulation of the reactivity of oryzacystatin-II. J. Agric. Food Chem. 55: 1762-1766.
Ordonez-Gutierrez, L., M. Martinez, I. Rubio-Somoza, I. Diaz, S. Mendez, and J.M. Alunda. 2009. Leishmania infantum: antiproliferative effect of recombinant plant cystatins on promastigotes and intracellular amastigotes estimated by direct counting and real-time PCR. Exp. Parasitol. 123: 341346.
Pernas, M., R. Sanchez-Monge, L. Omez, and G. Salcedo. 1998. A chestnut seed cystatin differentially effective against cysteine proteases from closely related pests. Plant Mol. Biol. 38: 1235-1242.
Polson, A. 1990. Isolation of IgY from the yolks of eggs by a chloroform polyethylene glycol procedure. Immunol. Invest. 19: 253-258.
Rassam, M. and W.A. Laing. 2004. Purification and characterization of phytocystatins from kiwifruit cortex and seeds. Phytochemistry 65: 19-30.
Shatters, R.G. Jr., M.G. Bausher, W.B. Hunter, J.X. Chaparro, P.M. Dang, R.P. Niedz, R.T. Mayer, T.G. McCollum, and X. Sinisterra. 2004. Putative protease inhibitor gene discovery and transcript profiling during fruit development and leaf damage in grapefruit (Citrus paradisi Macf). Gene 326: 77-86.
Shyu, D.J.H., C.L. Chyan, J.T.C. Tzen, and W.M. Chou. 2004.
Molecular cloning, expression, and functional characterization of a cystatin from pineapple stem. Biosci. Biotechnol. Biochem. 68: 1681-1689.
Solomon, M., B. Belengh, M. Delledonne, and A. Levine. 1999. The involvement of cysteine proteases and protease inhibitor genes in programmed cell death in plants. Plant Cell 11: 431-444.
Thompson, J.D., D.G. Higgins, and T.J. Gibson. 1994. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680
|
|