|
|
|
|
|
|
|
|
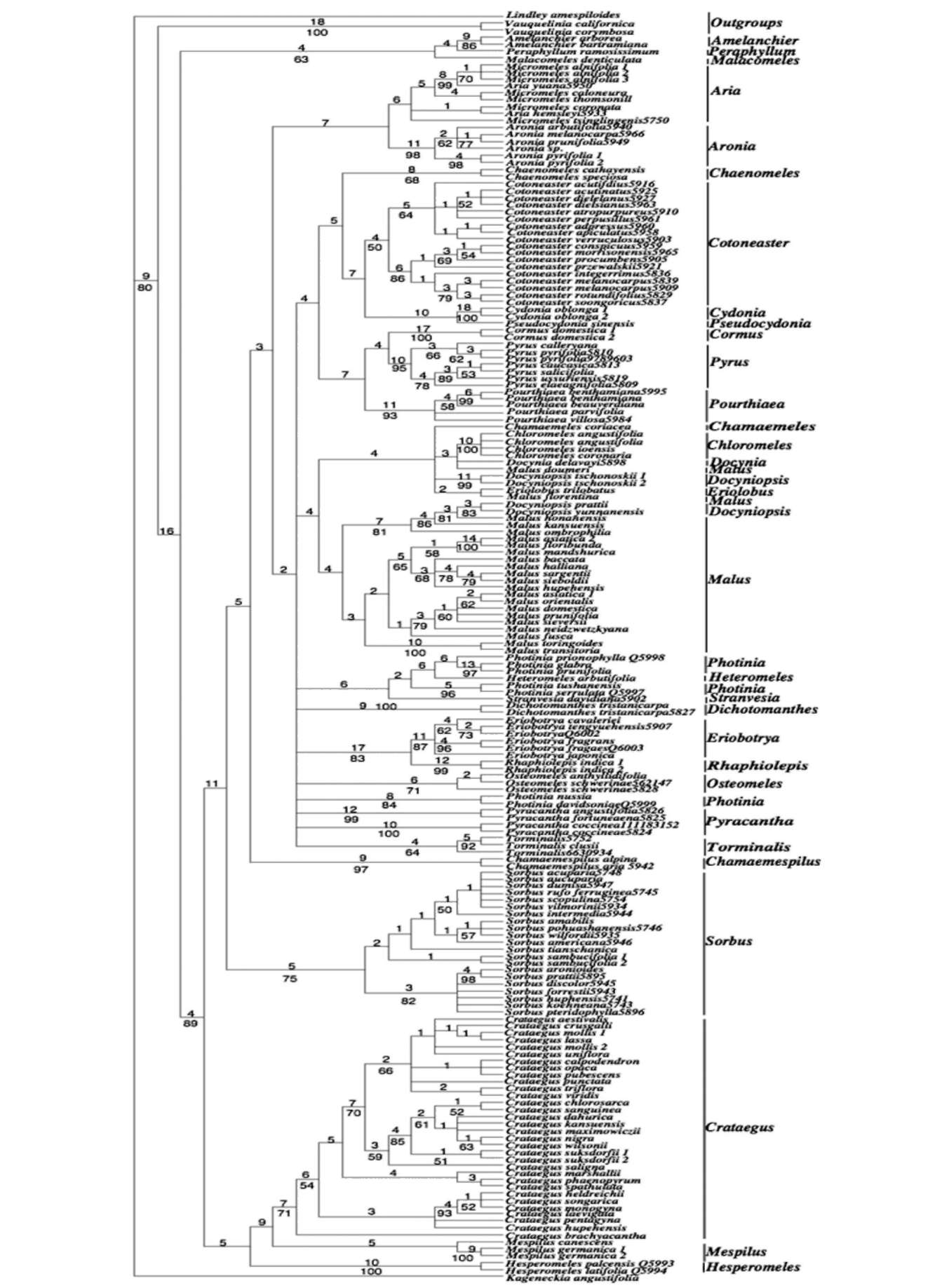
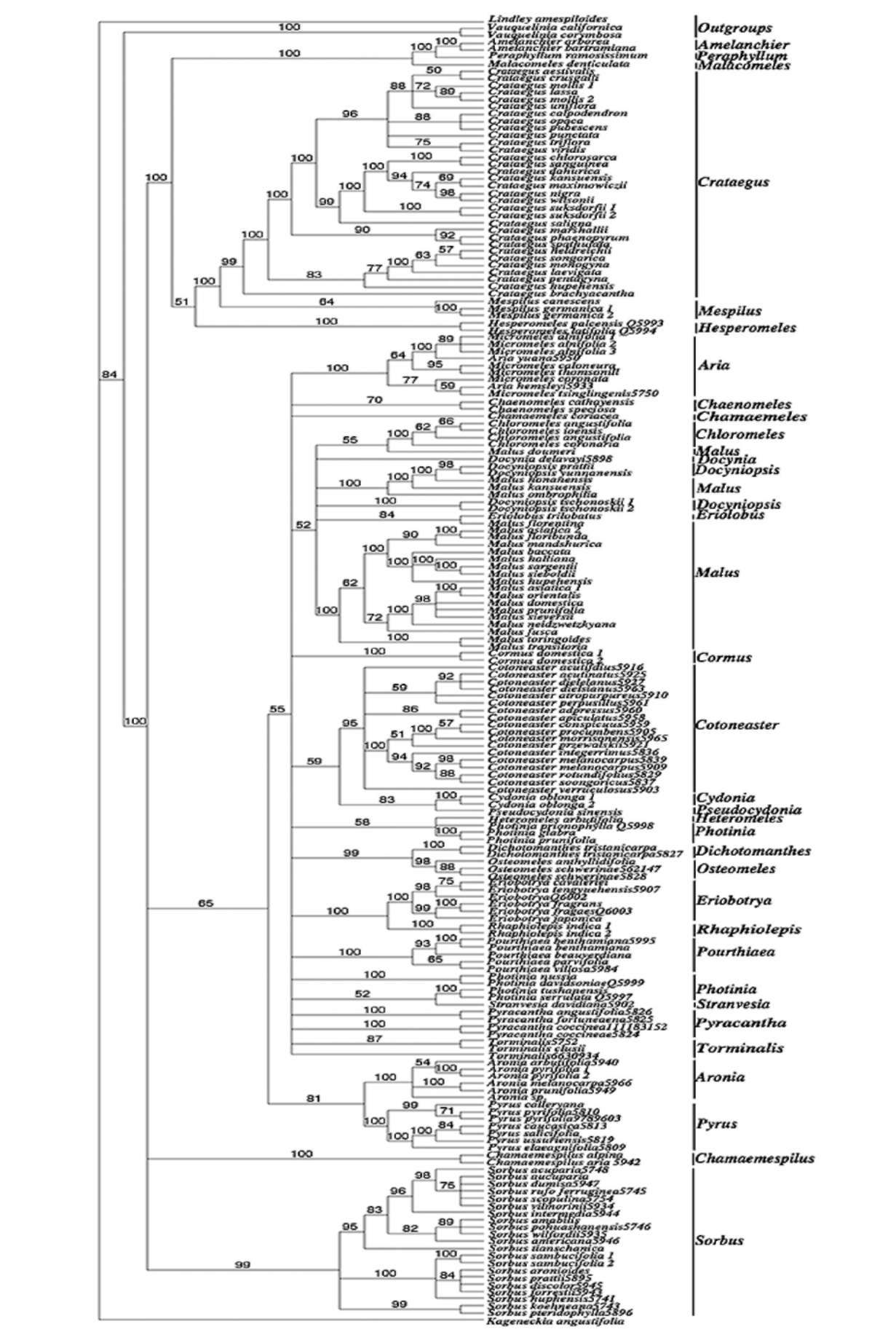
of Malus in having greenish, fragrant, often waxy fruits with a dense layer of sclereids around the core and just under the skin. Eriolobus, with a single species in the eastern Mediterranean, is unique in having deeply lobed simple leaves, incomplete adnation of carpels, and abundant sclereids in fruits. Docynia has two species, one in the Himalayas and from Assam to Vietnam and the other in southwestern China. Docyniopsis consists of four species, all in eastern Asia, and differs from Docynia in having only two ovules per locule (vs. 3-10 in Docynia). Nevertheless, the two genera share similar flavonoids chemistry (Williams, 1982). In the phylogenies, Docynia delavayi C.K. Schneid. is closely related to Malus doumeri A. Chev., M. florentina C.K. Schneid. and Eriolobus. Docyni-opsis tschonoskii (Maxim.) Koidz., D. prattii (C.K. Sch-neid.) Koidz., and D. yunnanensis (C.K. Schneid.) Koidz. do not form a clade, and the latter two species are closely allied with Malus honanensis Rehder, M. kansuensis (Batalin) C.K. Scheid., and M. ombrophila Hand.-Mazz. (bs=86%, pp=100%). Chloromeles forms a clade, but its relationship with other clades within Malus remains unresolved. Similarly, Docyniopsis, Eriolobus and Malus form a robust clade (95%) in Campbell et al.'s (2007) GBSSI-2B tree, but their relationships are unresolved. Therefore, it is appropriate to circumscribe Malus broadly, containing Chloromeles, Docynia, Docyniopsis, Eriolobus, and Malus.
|
|
|
|
Some authors recognize Sorbus in the broad sense, while others divide it into five genera (Robertson et al., 1991): Sorbus, Aria, Cormus, Torminalis, and Chamae-mespilus. A major reason that taxonomists in Europe and western Asia include these other genera in Sorbus is the large number of apomictic microspecies intermediate between them in those regions (McAllister H. 2005). Robertson et al. (1991) cited several examples of intergeneric hybrids involving Sorbus and other genera of the Pyrinae, such as xSorbocotoneaster, xSorbaronia, xAmelosorbus and x Crataegosorbus, and concluded that "the extensive hybridization between genera and subgeneric groups seems to reflect weak overall barriers to hybridization rather than indicating evolutionary relationships", and "it seems best to discount intergeneric hybridization when setting generic limits."
|
|
|
|
Cormus and Sorbus have pinnately compound leaves, Torminalis leaves are pinnately lobed, and those of Chamaemespilus are simple and toothed with campto-dromous venation. However, Aria is diverse in leaf morphology; some species have coarsely toothed leaves with craspedodromous venation, while others have simple leaves and camptodromous venation (Robertson, 1992). Kovanda and Challice (1981) segregated species with deciduous calyx lobes into Micromeles. However, the calyx feature is inconsistent in the Pyrinae, and thus Micromeles should not be recognized (Robertson, 1992; Rohrer et al., 1991). In the ITS trees, Micromeles species are intermixed with those of Aria (Figures 1-2), while Cormus, Tormina-lis, Chamaemespilus each form their own clades. Our ITS
|
|
|
|
|
|
|
|
|
|
|
|
|
data thus support their generic status in the Pyrinae.
|
|
|
|
|
|
|
|
Our ITS data, from multiple species representing the diversity of traditionally recognized genera, support recognition of 24 genera that are resolved as monophyl-etic: Amelanchier, Aria (including Micromeles), Aronia, Chaenomeles, Chamaemespilus, Chamaemeles, Cormus, Cotoneaster, Crataegus, Cydonia, Dichotomanthes, Eriobotrya, Hesperomeles, Malacomeles, Malus (including Chloromeles, Docynia, Docyniopsis, and Eriolobus), Mespilus, Osteomeles, Peraphyllum, Pourthiaea, Pseudo-cydonia, Pyrus, Rhaphiolepis, Sorbus, and Torminalis.
Most of these genera are essentially in agreement with recent works (Robertson et al., 1991). Among those genera, Aronia and Pourthiaea are separated from Photinia as independent genera, and Pourthiaea is for the first time supported by molecular data as a genus; Hesperomeles is also examined for the first time using molecular data and may have a close relationship to Crataegus-mespilus instead of Osteomeles. Our data support the inclusion in Malus of Chloromeles, Docynia, and Docyniopsis and suggest that Pyracantha may be polyphyletic. Photinia is found to be polyphyletic and possibly closely related to Heteromeles and Stranvaesia. However, more extensive sampling is needed to determine the generic limits of Py-racantha, Photinia, and Stranvaesia.
|
|
|
|
Acknowledgements. We thank Kyle Port, Kathryn Richardson and Eric Youngerman for their help in collecting plant material from the Arnold Arboretum, Jim Solomon of Missouri Botanical Garden for providing Hesperome-les leaf material, and Margaret Frank for lab assistance. Qingyan Li is grateful to the China Scholarship Council for a foreign study scholarship. This project was partially supported by grants from the National Natural Science Foundation of China (#30670141, #31170202) and the National Infrastructure of Natural Resources for Science and Technology (2005DKA21403) to Wenbo Liao.
|
|
|
|
|
|
|
|
Campbell, C.S., C.W. Greene, and T.A. Dickinson. 1991. Reproductive biology in subfam. Maloideae, Rosaceae. Syst. Bot. 16: 333-349.
|
|
|
|
Campbell, C.S., M.J. Donoghue, B.G. Baldwin, and M.F. Wojciechowski. 1995. Phylogenetic-Relationships in Maloideae (Rosaceae) - Evidence from Sequences of the In-ternal Transcribed Spacers of Nuclear Ribosomal DNA and Its Congruence with Morphology. Amer. J. Bot. 82: 903-918.
|
|
|
|
Campbell, C.S., M.F. Wojciechowski, B.G. Baldwin, L.A. Alice, and M.J. Donoghue. 1997. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Mol. Biol. Evol. 14: 81-90.
|
|
|
|
|
|
|
|