266
Botanical Studies, Vol. 53, 2012
on morphological similarities. Hence classifications disagree on how many species are present in East Asia, and molecular data provides a means of assessing the accuracy of the two taxonomic treatments.
Given the uncertainty regarding the number of taxo-nomic species in eastern Asian Nyssa, a DNA-based approach seems appropriate. Therefore, the objectives of this study are to: 1) test the efficiency of the four loci (matK, rbcL, trnH-psbA and ITS) as barcodes for Nyssaceae; and 2) use data from these to reevaluate the taxonomic issues.
MATERIALS AND METHODS
Plant materials
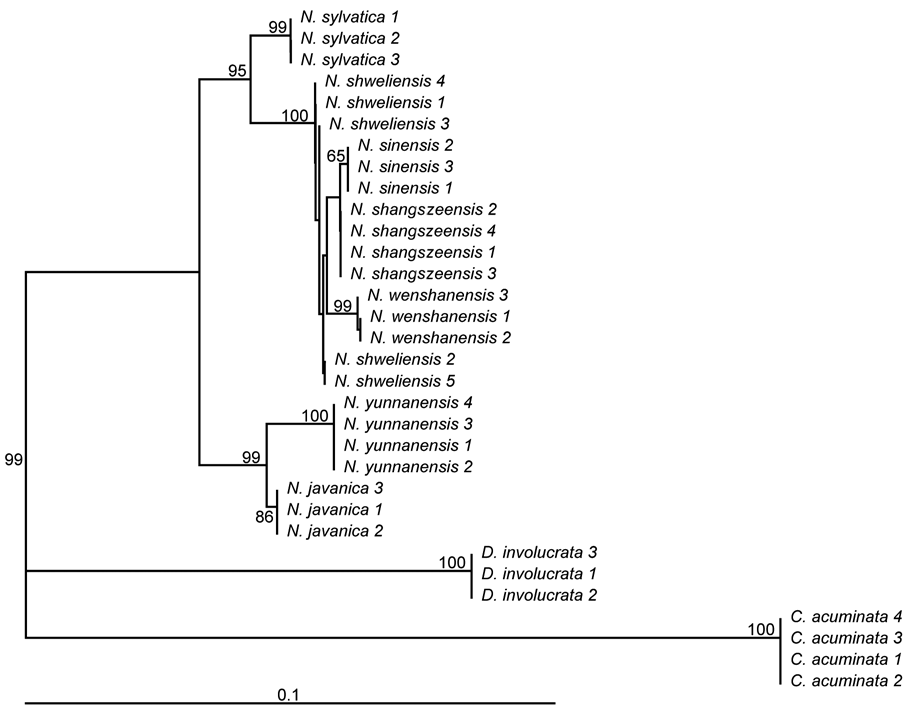
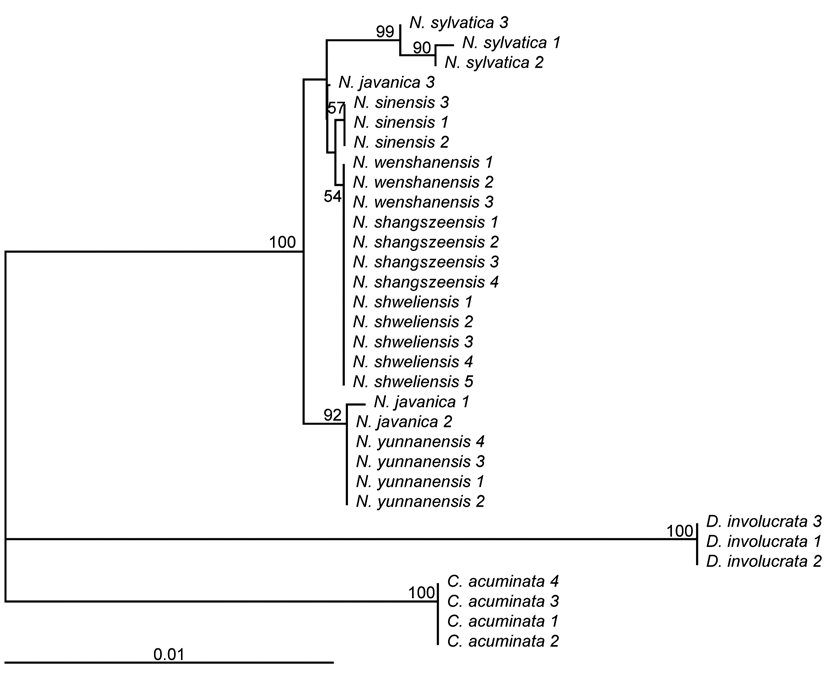
To gain a more comprehensive understanding, we sampled the species as recorded in Flora of China. Material of N. leptophylla Fang et Soong could not be obtained because insufficient information on its distribution (Sun, 2008) prevented us from locating it in the field. From the other six species, between three and five accessions were collected per species (Figure 1; Table S1). In addition, three accessions of one of the American species, N. sylvat-ica, were obtained from the Arnold Arboretum of Harvard University. Three and four accessions each of species from two other genera of Nyssaceae, Camptotheca acuminata and Davidia involucrata were also included (Table S1). Hence a total of 32 accessions from nine species were used to evaluate the candidate barcodes.
Material of N. yunnanensis and N. javanica was collected from one locality each (Table S1) and could be assigned to these species based on morphology. However, past taxo-nomic confusion combined with morphological similarity made identification of the other four species uncertain. Each of N. shangszeensis and N. wenshanensis have only ever been collected from their type localities (Table S1), and in each case no other Nyssa species has ever been recorded from the vicinity of their type locality. Our own
field observations confirmed the presence of a species matching the type description, and no other, at each type locality. Therefore, material from the type locality of each species was assigned to that species in each case. Similarly, material from a locality where N. sinensis (and no other species) and N. shweliensis (and no other species) had previously been recorded was examined, confirmed to match descriptions of each species, and therefore assigned to N. sinensis and N. shweliensis respectively. From each wild population of each species, each individual sampled was at least 30 meters from the nearest sampled accession. In addition, one cultivated accession of N. sinensis from Kunming was examined.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from 30-50 mg of silica-gel dried leaves of all accessions using a modified CTAB (cetyl trimethyl ammonium bromide) method (Doyle and Doyle, 1987). The extracted total DNA was diluted to a final concentration of 50-80 ng/[il for subsequent use. Am-plifications of the target gene regions were performed via the polymerase chain reaction (PCR) in a GeneAmp PCR System 9700 DNA Thermal Cycler (PerkinElmer, USA) or Eppendorf (Hamburg, Germany).
Nuclear ribosomal ITS and three chloroplast genes (matK, rbcL and psbA-trnH) were amplified (White et al. 1990; Tate and Simpson, 2003; Dunning and Savolainen, 2010; Ivanova et al., 2008). The PCR amplifications were carried out in 25 [il mixtures contained 0.3 U AmpliTaq polymerase, 10 x buffer, 1.5 mmol/L MgCl?, 0.2 mmol/L dNTP, 0.5 imol/L primer and 30-50 ng genomic DNA. The PCR conditions consisted of an initial denaturation at 95°C for 2-3 min, followed by 32-34 cycles of 1 min at 94°C, 50 sec at 52°C, 1.5 mm at 72°C, and ended with an extension step of 7 min at 72°C. The PCR products were purified using the GELase Agarose (Epicentre Technologies, Madison, WI, USA) according to the manufacturer's protocol for sequencing PCR reactions. Sequencing reactions were performed using PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). The sequencing products were run on an ABI 3700 automated sequencer (Perkin Elmer). All sequences were deposited in GenBank with the accession numbers listed in Table S1.
Data analysis
Sequences from each DNA region were aligned using the CLUSTAL X version 1.83 (Thompson et al., 1997) then refined manually. The inter- and intraspecific divergences of each barcoding region was computed by calculating Kimura 2-parameter (K2P) distances in MEGA 4 (Tamura et al., 2007). The significance of divergence was then assessed by Wilcoxon signed rank and Wilcoxon two-sample tests (http://www.fon.hum.uva.nl/Service/Statis-tics.html). The neighbor-joining (NJ) method was used to construct phylogenetic trees in PAUP 4.0b10 (Swofford et
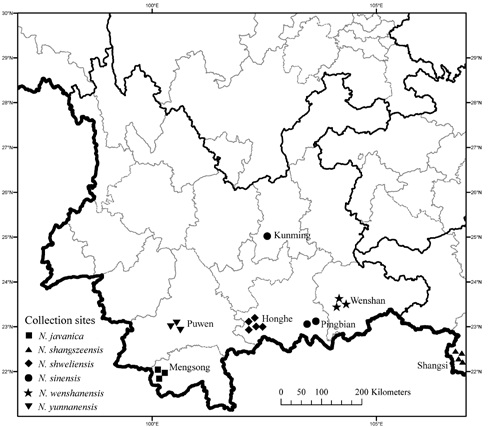
Figure 1. The localities of Nyssa species used in this study.