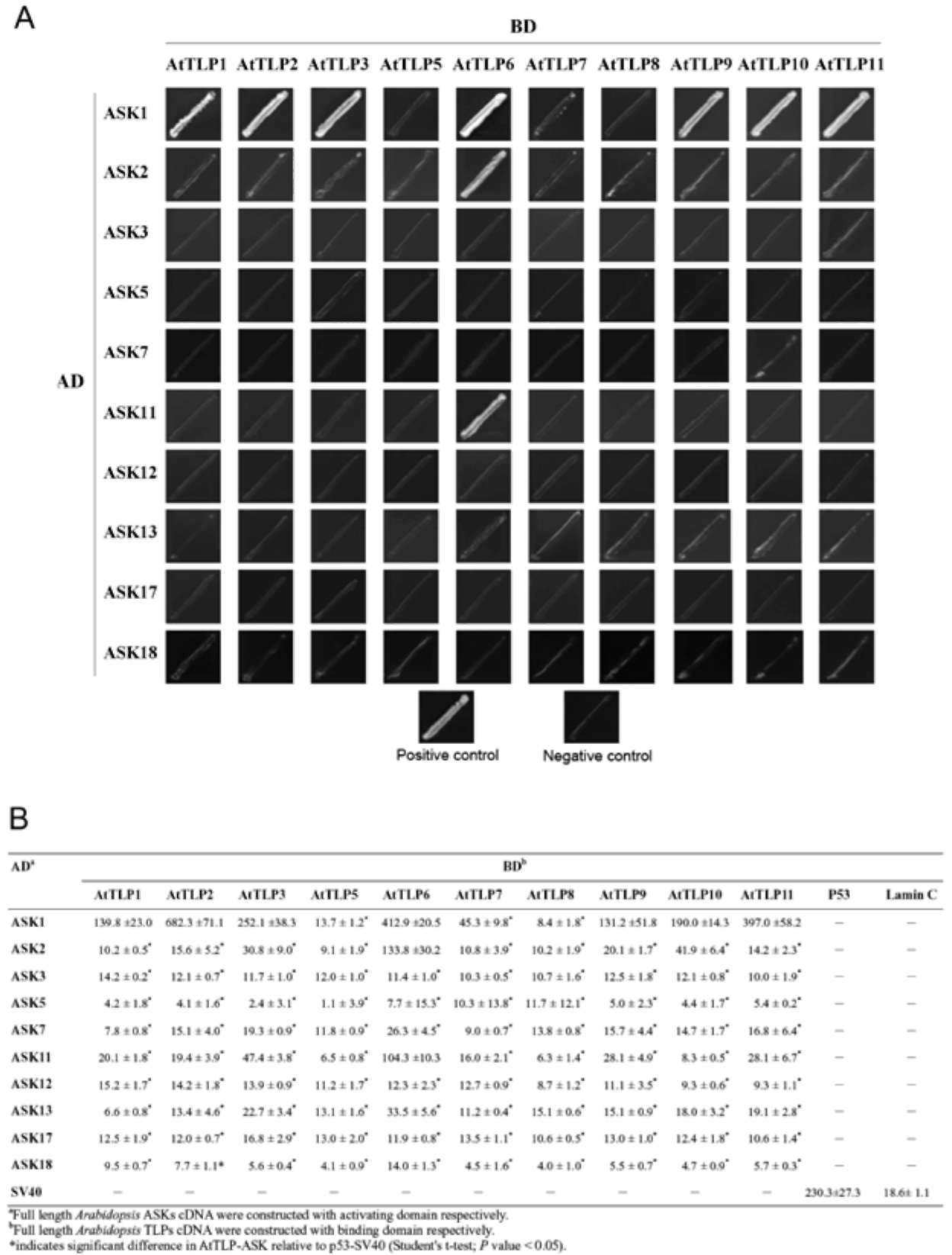
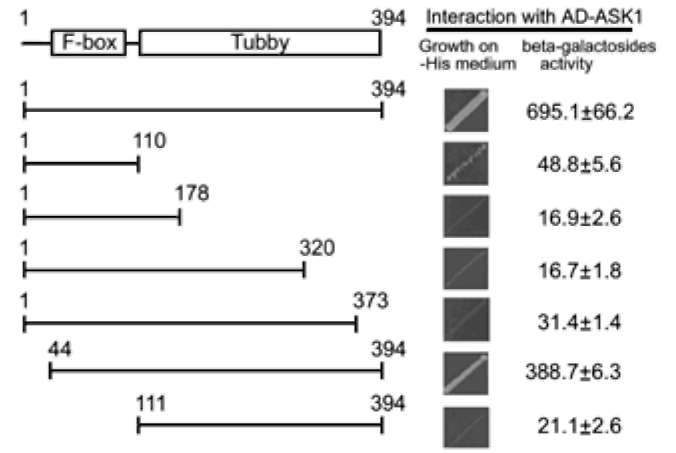
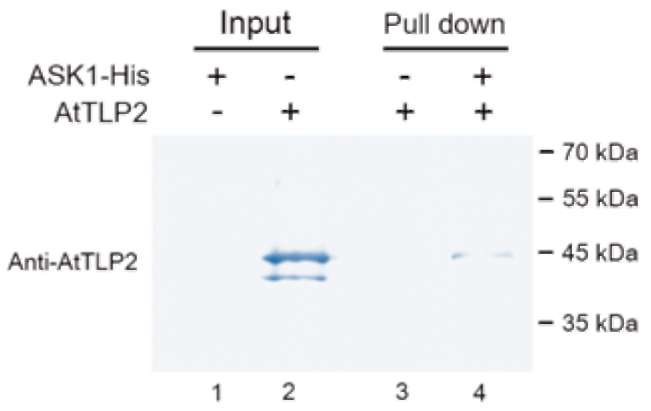
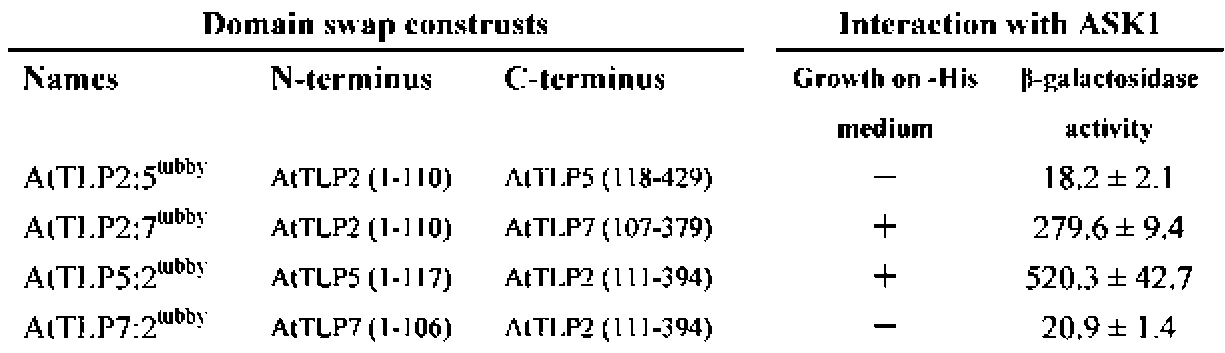
E.A Woolf, O. Tayber, T. Brody, P. Shu, F. Hawkins, B. Kennedy, L. Baldini, C. Ebeling, G.D. Alperin, J. Deeds, N.D. Lakey, J. Culpepper, H. Chen, M.A. Glucksmann-Kuis, G.A. Carlson, and G.M. Duyk, and K.J. Moore. 1996. Identification and Characterization of the Mouse Obesity Gene tubby: A Member of a Novel Gene Family. Cell 85:281-290.
Lai, C.P., C.L. Lee, C. P.H. Chen, S.H. Wu, C.C. Yang, and J.F. Shaw. 2004. Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 134: 1586-1597.
Lai, C.P., P.H. Chen, J.P. Huang, Y.H. Tzeng, S.M. Chaw, J.F. Shaw. 2012. Functional diversification of the tubby-like protein gene families (TULPs) during eukaryotic Evolution. Biocatal. Agri. Biotechnol. 1: 2-8.
Li, Q.Z., C.Y. Wang, J.D. Shi, Q.G. Ruan, S. Eckenrode, A. Davoodi-Semiromi, T. Kukar, Y. Gu, W. Lian, D. Wu, and J.X. She. 2001. Molecular cloning and characterization of the mouse and human TUSP gene, a novel member of the tubby superfamily. Gene 273: 275-284.
Mukhopadhyay, S., X. Wen, B. Chih, C.D. Nelson, W.S. Lane, S.J. Scales, and P.K. Jackson. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24: 2180-2193.
Nishina, P. M., M.A. North, A. Ikeda, Y. Yan, and J.K. Naggert. 1998. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics
54: 215-220.
Noben-Trauth, K., J.K. Naggert, M.A. North, and P.M. Nishina. 1996. A candidate gene for the mouse mutation tubby. Nature 380: 534-538.
North, M. A., J.K. Naggert, Y. Yan, K. Noben-Trauth, and P.M. Nishina. 1997. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc. Nat. Acad. Sci. 94: 3128-3133.
Patton, E.E., A.R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin- dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14: 236-243.
Risseeuw, E.P., T.E. Daskalchuk, T.W. Banks, E. Liu, J. Cote-lesage, H. Hellmann, M. Estelle, D.E. Somers, and W.L. Crosby. 2003. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34: 753-767.
Russell, I.D., A.S. Grancell, and P.K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the
CBF3 kinetochore complex. J. Cell Biol. 145: 933-950.
Samach, A., J.E. Klenz, S.E. Kohalmi, E. Risseeuw, G.W. Haughn, and W.L. Crosby. 1999. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral mer-istem, Plant J. 20: 433-445.
Santagata, S.T.J., Boggon, C.L. Baird, J.A. Gomez, J. Zhao, W.S. Shan, D.G. Myszka, and S.L. Shapiro. 2001. G-protein signaling through tubby proteins. Science. 292: 2041-2050.
Schulman, B., A. A.C. Carrano, P.D. Jeffrey, Z. Bowen, E.R.