|
|
|||||
|
Botanical Studies (2009) 50: 435-442.
|
PHYLOGENETICS
|
|
|||
|
|
|||||
|
|
|||||
|
Strong incongruence between the ITS phylogeny and generic delimitation in the Nemosenecio-Sinosenecio-Tephroseris assemblage (Asteraceae: Senecioneae)
Liu-Yang WANG1'2'5, Pieter B. PELSER3, Bertil NORDENSTAM4, and Jian-Quan LIU2 *
1Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Plateau Institute of Biology, Chinese Academy of Sciences, Xining 810001, PR. China
2Key Laboratory of Arid and Grassland Ecology, School of Life Science, Lanzhou University, Lanzhou 730000, PR. China 3Oklahoma State University, Botany Department, 104 Life Sciences East Stillwater, Oklahoma 74078-3013, USA 4Swedish Museum of Natural History, P. O. Box 50007, SE-104 05 Stockholm, Sweden 5 Graduate University of Chinese Academy of Sciences, Beijing 100049, PR. China
(Received January 9, 2009; Accepted March 9, 2009)
ABSTRACT. The three genera Sinosenecio, Nemosenecio and Tephroseris form a closely knit group nested in the subtribe Tussilagininae of the tribe Senecioneae (Asteraceae). The generic limits in this assemblage remain unclear and need revision. In this study, we analysed sequences of the internal transcribed spacer (ITS) region of nuclear ribosomal DNA available from GenBank and sequenced 19 accessions of an additional 13 species encompassing all three genera. Phylogenetic analyses based on the ITS variation of 27 species in this assemblage and seven species from related genera of the Tussilagininae suggested that neither Sinosenecio nor Tephroseris is monophyletic. The sampled species of Sinosenecio were scattered in different clades or subclades of the phylogenetic tree. Four species of this genus, including the generic type species (S. eriopodus, S. hederifolius, S. homogyniphyllus and S. subcoriaceus) are clustered in a tentative clade with genera such as Ligularia, Cremanthodium, Parasenecio, Farfugium and Tussilago. The remaining ten Sinosenecio species comprise a highly supported clade together with 13 Tephroseris species and four Nemosenecio species. Within this clade, 10 Tephroseris species together with two Sinosenecio species (S. ne^wcombei and S. koreanus) comprise a monophyletic subclade while the remaining 11 species from all of three genera are clustered into another clade with moderate statistical support. Within the latter subclade, T. changii was revealed to be closely related to four Sinosenecio species, and three Nemosenecio species comprising a monophyletic lineage. These two lineages form a polytomous radiation with the other two Sinosenecio lineages. The generic delimitations of the three genera clearly need some adjustments, which is also supported by previous studies of gross and floral morphology. Two Sinosenecio species (S. newcombei and S. koreanus) should be transferred to Tephroseris, and the genus Sinosenecio should be re-circumscribed to contain those species clustered in the Ligularia―Tussilago clade. Most of the other described species under Sinosenecio and T. changii should either be transferred to an enlarged Nemosenecio concept, or a new genus needs to be established to encompass them. However, the morphological distinctions between these genera require further investigation.
Keywords: Asteraceae; Internal transcribed spacer (ITS) region; Nemosenecio; Molecular phylogeny; Sinosenecio; Tephroseris; Tussilagininae.
|
|||||
|
|
|||||
|
INTRODUCTION
Nemosenecio (Kitam.) B. Nord., Sinosenecio B. Nord. and Tephroseris (Reichenb.) Reichenb. are closely related genera of tribe Senecioneae of the Asteraceae (Liu et al., 2006; Pelser et al., 2007). Nemosenecio has six species, five of which are found in China and one in Japan (Jeffrey and Chen, 1984; Nordenstam, 2007; Zhang
|
et al., 2008). Sinosenecio contains about 38 species of which 37 occur in China, Korea, and Indo-China with a distinct center of diversity in the Sichuan province of China. Sinosenecio ne^wcombei (Greene) J. P. Janovec & T. M. Barkley, however, is endemic to the Queen Charlotte Islands of Canada. Tephroseris contains around 50 species mainly found in temperate and arctic Eurasia. Six species are recognized in NW North America, one of which is endemic (Barkley and Murray, 2006). Although these three genera have traditionally been regarded as members of the subtribe Tussilagininae Dum., the Nemosenecio-Sinosenecio-Tephroseris assemblage has
|
||||
|
*Corresponding authors: E-mail: liujq@nwipb.ac.cn; ljqdxy@public.xn.qh.cn; Fax: +86-931-8914288; Tel: +86-931-8914305.
|
|||||
|
|
|||||
|
|
|||
|
436
|
Botanical Studies, Vol. 50, 2009
|
||
|
|
|||
|
also been recognized at the subtribal level (Jeffrey and Chen, 1984). Tephroseridinae C. Jeffrey et Y. L. Chen was considered as distinct from the Tussilagininae by owning narrow, cylindrical anther-collars, polarized, scattered, or radial endothecial cell wall thickenings, and confluent, contiguous or separate stigmatic areas (Jeffrey and Chen, 1984). These character states are, however, certainly not unique to Nemosenecio, Sinosenecio, and Tephroseris and have also been observed in the Tussilagininae sensu Jeffrey and Chen (Liu, 1999). In addition, Jeffrey and Chen (1984) suggested the gametic chromosome number of 24 as diagnostic for Tephroseridinae, although a wide range of other chromosome numbers have also been observed (Jeffrey and Chen, 1984; Nordenstam, 2007; Pelser et al., 2007) and the more typical Tussilaginoid chromosome number of x = 30 has been recorded for two Sinosenecio species (Liu, 2004). Of the three genera, Sinosenecio especially shows character states of both the Tephroseridinae and Tussilagininae sensu Jeffrey and
Chen (1984) (Liu, 2000). Because of the lack of diagnostic
characters for Tephroseridinae and its phylogenetic position deeply nested within Tussilagininae sensu Jeffrey and Chen (1 984), it is currently not recognized as a subtribe and Nemosenecio, Sinosenecio, and Tephroseris are placed in Tussilagininae by most authors (e.g., Bremer,
1994; Liu et al., 2006; Pelser et al., 2007).
In addition to difficulties concerning the subtribal delimitation, also the generic delimitation of the Nemosenecio-Sinosenecio-Tephroseris assemblage has been somewhat problematic. The three genera have mainly been defined on the basis of leaf characters
(Jeffrey and Chen, 1984), although Nordenstam (1978)
also indicated differences in habit and floral morphology. Both Nemosenecio and Tephroseris have pinnately-veined leaves, but those of Nemosenecio are pinnatisect, whereas the leaves of Tephroseris are subentire or only shallowly lobed. In contrast to these two genera, most species of Sinosenecio have palmately-veined leaves with sinuate-dentate to sinuate-denticulate margins, although the leaves of Sinosenecio hainanensis (Chang & Tseng) C. Jeffrey & Y. L. Chen are pinnately-veined. Although Nemosenecio is easily distinguished by its deeply incised leaves, the differences between Sinosenecio and Tephroseris are not always clear. Tephroseris changii B. Nord, for example, was regarded as a member of Tephroseris based on its pinnately veined leaves, even though this species resembles some of the Sinosenecio species in habit, anther shape and phyllary number (Jeffrey and Chen, 1984).
In recent years, phylogenetic studies of DNA sequences have been successfully applied to resolve the systematic positions and generic delimitations of several Senecioneae genera (e. g. , Knox and Palmer, 1 99 5; S wenson and
Bremer, 1997, 1999; Panero et al., 1999; Bain and Golden, 2000; Wagstaff and Breitwieser, 2004; Liu et
al., 2006; Pelser et al., 2002, 2003, 2007; Wagstaff et al.: 2006). These molecular phylogenetic studies suggested that the morphology-based generic delimitation of the
Nemosenecio-Sinosenecio-Tephroseris assemblage needs
|
to be revised to resolve monophyletic genera. For example, Golden et al. (2001), prompted by the extraordinary biogeographical consequence of the transfer of Senecio newcombei Greene from the Queen Charlotte Islands to the otherwise largely Chinese Sinosenecio; Janovec and Barkley (1996) performed a phylogenetic study of ITS sequences to examine the true relationships of S. newcombei. These authors found that the two species of Sinosenecio included in their studies (S. koreanus (Kom.) B. Nord. from Korea and S. newcombei) did not form a monophyletic group and are nested within Tephroseris. The non-monophyly of Sinosenecio was also concluded in another study using ITS sequence data (Liu et al., 2006). In that study, the single species of Tephroseris and Nemosenecio and two of the three Sinosenecio species included formed a well supported clade in which S. bodinieri (Van.) B. Nord. and S. globigerus (Chang) B. Nord. comprised a strongly supported subclade. Sinosenecio subcoriaceus C. Jeffrey & Y. L. Chen, the third species included, however, appeared to be more distantly related and was placed in a large polytomy with several other Tussilagininae clades. The results of these two studies were confirmed by Pelser et al. (2007), who included the Nemosenecio, Sinosenecio, and Tephroseris ITS sequences generated by Golden et al. (2001) and Liu et al. (2006) and a few other accessions of Sinosenecio and Tephroseris in their Senecioneae ITS phylogeny. This study indicated with strong bootstrap support and posterior probabilities that Sinosenecio newcombei and S. koreanus are more closely related to Tephroseris than to the other four Sinosenecio species that were included. Furthermore, Tephroseris changii proved to be distantly related to the other Tephroseris species and instead formed a clade with Sinosenecio bodinieri, S. globigerus, and S. septilobus (Chang) B. Nord. In the same way as Liu et al. (2006), Pelser et al. (2007) found S. subcoriaceus to be only distantly related to the other members of the Nemosenecio-Sinosenecio-Tephroseris assemblage and nested within a large, but poorly resolved clade composed of Cremanthodium, Ligularia, Parasenecio, and other mostly Asian Tussilagininae genera.
As a first step towards arriving at a monophyletic generic delimitation of the Nemosenecio-Sinosenecio-Tephroseris assemblage, an ITS phylogeny of this group is presented that includes a much larger sampling of its species (27) together with a selection of other Senecioneae genera. On the basis of this phylogeny, we discuss its incongruence with the current morphology-based generic delimitation and explore alternative classifications to obtain strictly monophyletic genera.
MATERIALS AND METHODS
Sequence data and sampled species
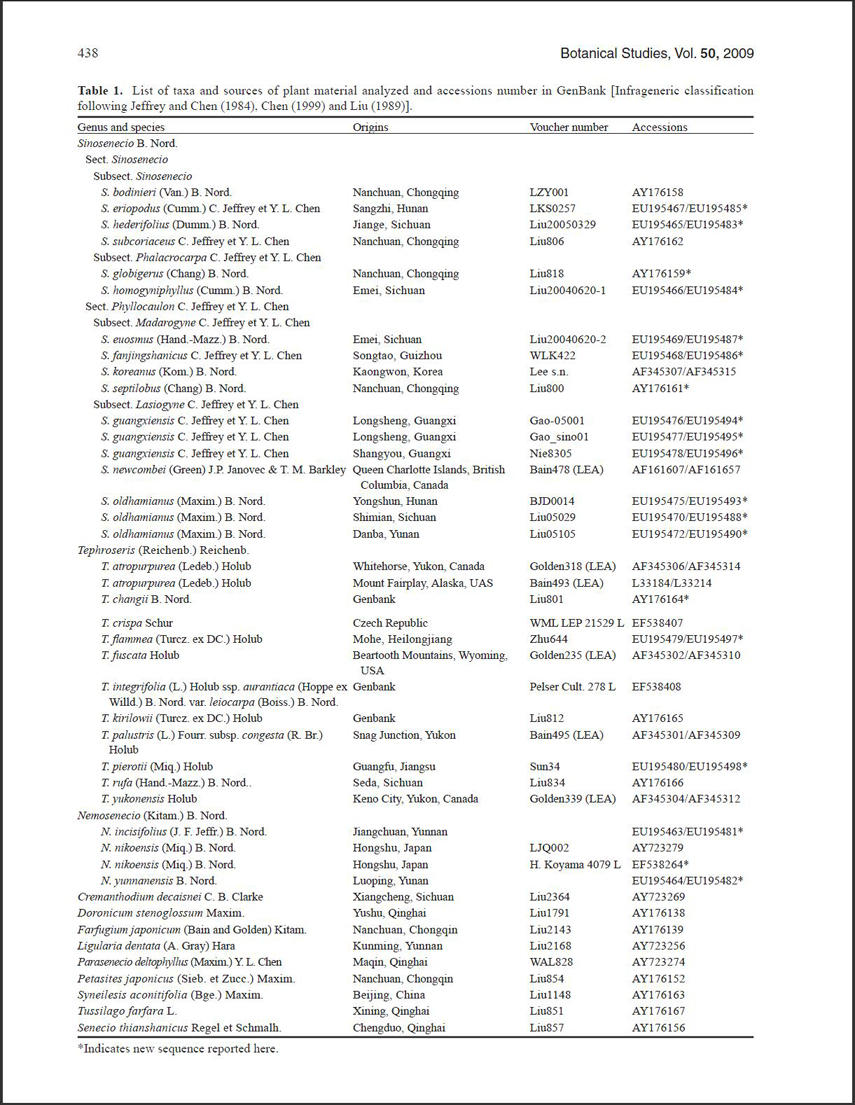
The ITS sequences included in this study were either newly obtained (19 accessions representing 13 species) or downloaded from GenBank. Voucher information and
|
||
|
|
|||
|
|
|||
|
WANG et al. ― Recircumscription of subtribe Tephroseridinae
|
437
|
||
|
|
|||
|
GenBank accession numbers are listed in Table 1. In total, our data set comprises 42 accessions of 27 species (Table 1) representing three Nemosenecio species, 11 Tephroseris species listed by Jeffrey & Chen (1984), and 13 Sinosenecio species selected to represent the two sections, four subsections, and four series that Jeffrey & Chen (1984) recognized for Sinosenecio (Table 1). Our samples comprised all sections and subsections of Sinosenecio, and species which have represented the distribution range of Tephroseris (see details to Table 1). In addition, seven species from seven other Tussilagininae genera were included. Subtribe Senecioninae was represented with a single species (Senecio thianshanicus Regel et Schmalh.) and Doronicum stenoglossum Maxim. was selected as outgroup on the basis of the results of previous studies (Liu
et al., 2006).
DNA extraction, amplification and sequencing
Total DNA was extracted from fresh or silica-gel dried leaf tissue or from leaf samples taken from herbarium specimens using the DNeasy Plant Mini kit (QIAGEN, Valencia, CA) following the manufacturer's protocol. ITS was amplified with primers "1a" and "4" (White et al., 1990). Polymerase chain reactions (PCR) were performed in a 25-[il volume, containing 10-40 ng plant DNA, 50 mM Tns-HCI, 1.5 mM MgCl^ 250 ^g/mL BSA, 0.5 mM dNTPs, 2 fiM of each primer, and 0.75 unit of Taq polymerase. PCR reactions were performed with the following thermocycling conditions: 5 min at 95°C, 36 cycles of 1 min at 94°C, 1 min of annealing at 52°C, and 1.25 min at 72°C, with a final 8 min extension at 72°C, and reactions were kept at 4°C until further processing. PCR products were purified using a TIANquick Midi Purification Kit following the recommended protocol (TIANGEN). Sequencing reactions were performed with the PCR primers using the ABI Prism BigdyeTM Terminator Cycle Sequencing Ready Reaction Kit. Both the forward and reverse strands of DNA were sequenced and this resulted in a minimum overlap of 70% of their length in the contigs. Sequences were aligned using CLUSTAL X (Thompson et al., 1997) with default parameter settings and were edited by hand. The boundaries of the ITS region were determined by comparison with the results of Liu et al. (2006). All new sequences have been deposited in GenBank under accession numbers EU195463-EU195532 (Table 1).
Data analysis
Phylogenetic trees were reconstructed from ITS sequences with maximum likelihood (ML) and maximum parsimony (MP) using PAUP* v 4.0b10 (Swofford, 2002) and Bayesian Inference (BI) with MrBayes 3.0 (Huelsenbeck and Ronquist, 2001; Huelsenbeck et al.,
2001; Ronquist et al., 2003).
MP analyses involved a heuristic search strategy with 100 replicates of random addition of sequences, in combination with ACCTRAN character optimization,
|
MULPARS + TBR branch swapping and STEEPEST DESCENT options on. Bootstrap values (BS; Felsenstein, 1 985) were calculated from 1000 replicates using a heuristic search with simple addition with TBR and
MULPARS options on.
For the ML analyses, an appropriate nucleotide substitution model was selected using the Akaike Information Criterion (AIC) implemented in MODELTEST version 3.06 (Posada and Crandall, 1998), and a heuristic search with simple addition of sequences
and TBR branch swapping, MULTREES and COLLAPSE
was used to produce ML trees.
The nucleotide substitution model selected with MODELTEST was also used for the BI analyses, which were carried out with four simultaneous Monte-Carlo Markov Chains (MCMC; three heated and one cold) run for two million generations. Trees were saved every 100 generations. A burn-in of 5000 trees was discarded after visual inspection of the log-likelihood values and the remaining 15 001 trees were used to construct a 50% majority rule consensus tree with posterior probabilities
(PP).
RESULTS
The aligned ITS matrix contained 592 characters, of
which 283 (47.80%) were constant and 172 (15.72%) were
parsimony-informative. The best-fit model and parameters
selected for this data set by the AIC in MODELTEST
were: GTR + G; base = (0.2386, 0.2181, 0.2418), nst = 6, rmat = (0.7879, 2.0385, 1.2710, 0.5508, 3.5878), rates = gamma and shape = 1.2580.
MP, ML, and BI analyses resulted in trees with almost identical topologies, except for a few species that were placed in slightly different phylogenetic positions and received low BS and PP values in the MP and BI analyses. Only the ML tree is shown here (Figure 1). Two main clades were resolved for Tussilagininae, but only Clade A composed of Nemosenecio, Tephroseris and the majority of the Sinosenecio species, received strong BS support (99%) and PP (1.00). Four Sinosenecio species (S. eriopodus, S. hederifolius, S. homogyniphyllus and S. subcoriaceus), however, were not found to be part of Clade A and instead formed a poorly supported (BS < 50%; PP < 0.50) clade (B) together with Ligularia, Cremanthodium and the other Tussilagininae genera. Within clade A, one subclade (C) comprised 11 Tephroseris species and two Sinosenecio species (S. newcombei and S. koreanus) and received
high BS (100%) and PP (1.00). The other subclade (D)
contained 11 species from all three genera and was weakly
supported (BS of 70%; PP of 0.55). Within subclade D,
the three accessions of S. guangxiensis composed a clade sister to the remainder of D. Tephroseris changii, the only Tephroseris species of D, was found to be nested within a clade of four Sinosenecio species. All three species of Nemosenecio included in our studies comprise a monophyletic lineage within D.
|
||
|
|
|||
|
|
|||||
 |
|||||
|
|
|||||
|
|
||||||||||||
|
WANG et al. ― Recircumscription of subtribe Tephroseridinae
|
439
|
|||||||||||
|
|
||||||||||||
|
DISCUSSION
Our phylogenetic analyses confirm the results from previous ITS studies that used a much smaller sampling of the Nemosenecio-Sinosenecio-Tephroseris assemblage
(Golden et al., 2001; Liu et al., 2006; Pelser et al., 2007),
showing that neither Sinosenecio nor Tephroseris is monophyletic. The results of this study further indicate that Nemosenecio is monophyletic and deeply nested within a clade composed of the majority of Sinosenecio species included (Clade D). These findings have been confirmed also by preliminary phylogenetic analyses of a trnL-trnF data set that contains a selection of the Nemosenecio, Sinosenecio, and Tephroseris species included in the present study (unpublished data) and that will be expanded in future studies. Because of the incongruence between the molecular phylogenies and the current generic classification of the Nemosenecio-Sinosenecio-Tephroseris assemblage, its generic delimitation needs to be revised.
The re-circumscription of Tephroseris
All sampled Tephroseris species (10/11) except for T. changii constitute a well supported monophyletic lineage together with two Sinosenecio species (S. newcombei and
|
S. koreanus) (Figure 1). Because the Tephroseris species not included in this study have a habit and morphology that is similar with the species sampled here, it is highly likely that most species described under Tephroseris will be part of this lineage. Therefore, a newly circumscribed Tephroseris should contain most species previously placed in this genus (Jeffrey and Chen, 1984; Jeffrey, 1992) and the two Sinosenecio species.
Sinosenecio koreanus has a small distribution area in north Korea and the adjacent part of Jilin in NE China, far outside the center of diversity of Sinosenecio. Although its habit is Sinosenecio-like, the leaf-blades are not distinctly cordate as is common in Sinosenecio, but rather subtruncate to cuneate and not distinctly palmately veined; thus in these characters it more closely resembles Tephroseris. The petioles are not clearly winged like in many Tephroseris species, but they are at least basally expanded. The ray-florets of S. koreanus exceed the phyllaries in number (c. 18 and c. 13, resp.), another character unusual in Sinosenecio, where phyllaries usually equal or exceed rays in number.
The phylogenetic affinities of S. newcombei have long been unclear and this species has previously been included in Senecio (Greene, 1897) and Packera (Weber and Love,
|
|||||||||||
 |
Figure 1. The single ML tree based on ITS sequences for the Sinosenecio-Nemosenecio-Tephroseris assemblage and other genera of the subtribe Tussilagininae of the tribe Senecioneae. Letters A and B next to the bars represent for two major clades of the ML tree, while C and D stand for the two sub-clade of Nemosenecio-Sinosenecio-Tephroseris assemblage (for details, please refer the Result and Discussion part). Bootstrap values from the most parsimony analyses with 1000 replicates (above branch) and Bayesian posterior probabilities appear at branch nodes (under branch).
|
|||||||||||
|
|
||||||||||||
|
|
|||
|
440
|
Botanical Studies, Vol. 50, 2009
|
||
|
|
|||
|
1981) from where it was placed in Sinosenecio (Janovec and Barkley, 1996) to now eventually find a home in Tephroseris. Sinosenecio newcombei has a chromosome number of n = 24 (Taylor and Mulligan, 1968), which is characteristic of Tephroseris, although also found in Sinosenecio. On overall morphological features S. newcombei is better positioned in Tephroseris than in Sinosenecio.
The newly defined Tephroseris does not have morphological characters that allow easy distinction from Sinosenecio. Its species have leafy stems, but these are also found in Sinosenecio and the basic chromosome number of x = 24 or 12 (Liu, 2004) found in Tephroseris is also observed in Sinosenecio. Although most Tephroseris species have pinnately-veined leaves, these are palmately-veined in S. newcombei and S. koreanus. Anther-collars are cylindrical and endothecial cell wall thickenings are mainly polar although a few cells close to the connective tissue bear radial thickenings as well (Jeffrey and Chen,
1984; Golden et al., 2001; Liu, 2001) as is also the case
in the other genera of the Tusslagininae (Liu, 1999). The petioles of all these species are indistinct from the lamina
(Jeffrey and Chen, 1984; Golden et al., 2001), a feature
characteristic of Tephroseris.
The polyphyly of Sinosenecio
The ITS phylogenies (Figure 1 ) all indicate that Sinosenecio is polyphyletic and remains an unnatural group even if S. koreanus and S. newcombei are transferred to Tephroseris. Four of the Sinosenecio species included in our studies (S. eriopodus, S. hederifolius, S. homogyniphyllus, and S. subcoriaceus) form a clade with Cremanthodium, Farfugium, Ligularia, Parasenecio, Petasites, Syneilesis, and Tussilago (Clade B). This clade is sister to Clade A, which includes Nemosenecio, Tephroseris, and the other Sinosenecio species (Figure 1). As observed in other studies (Liu et al., 2006; Pelser et al., 2007) resolution in Clade B is poor and relationships do not conform well to the current generic delimitation. This is also found for the four Sinosenecio species in Clade B of which only S. homogyniphyllus and S. subcoriaceus form a clade. These two species are morphologically similar and no doubt closely related, and it should be noted that the former is the generic type. Although Clade B lacks diagnostic morphological characters, many of its species have a basic chromosome number of n = 30 (Liu, 2004) which is also found in S. hederifolius and S. subcoriaceus (Liu, 1999 and unpublished data). Reports of chromosome counts for S. eriopodus are not known to us, but Liu (1999) reported 2n = 24 from roots of S. homogyniphyllus. This finding, however, may need confirmation, because it is quite different from other counts for members of Clade B. Just like Farfugium, another genus in Clade B, all four Sinosenecio members of this clade have young leaves with involute leaf margins. Other Sinosenecio species with involute leaf margins, such as S. cyclaminifolius (Franch.) B. Nord. and S. dryas (Dunn) C. Jeffrey et Y. L. Chen,
|
may be closely related to these four Sinosenecio species, although this needs to be confirmed in future molecular and morphological studies. Because S. homogyniphyllus is the type species of the genus (Nordenstam, 1978), Sinosenecio has to be more narrowly defined to include only those species that are part of Clade B, or even a selection of them if more detailed studies indicate that these species do not form a monophyletic group.
The remaining seven Sinosenecio species included are members of subclade D, in which Tephroseris changii and Nemosenecio take nested positions (Figure 1). It is not surprising that T. changii appears to be closely related to S. septilobus, S. bodinieri, S. fangjingshanicus, and S. globigerus, because all of these species are scapigerous and have similar habit, anther shape and phyllary number (Jeffrey and Chen, 1984). Subclade D is characterized by scattered or radial endothecial cell wall thickenings
(Jeffrey and Chen, 1984; Liu, 2001), although these
character states are also found elsewhere in Tussilagininae, and chromosome numbers of 2n = 24, 48 or 72 (Liu, 1999, 2004). Because of their distant relationship with the type of Sinosenecio (S. homogyniphyllus), Tephroseris changii and the species of Sinosenecio in sublade D need to be accommodated in another genus, separate from Sinosenecio.s.s.. This could be achieved by transferring the seven Sinosenecio species and T. changii to Nemosenecio. Alternatively, in order to preserve Nemosenecio in its current circumscription, the clade composed of S. septilobus, S. bodinieri, S. fangjingshanicus, S. globigerus, and T. changii could be described as a new genus as well as the remainder of the subclades found in subclade D. In our opinion, however, subclade D is currently too poorly resolved to follow the latter taxonomic option and requires more detailed molecular and morphological studies before taxonomic changes should be made.
In conclusion, our studies indicate that the morphology-based generic delimitation of the Nemosenecio-Sinosenecio-Tephroseris assemblage is strongly incongruent with the ITS phylogeny. This most likely means that the morphological characters used to define genera (e.g., leaf venation) show widespread homoplasy. Future morphological and cytological studies could reveal new diagnostic characters for clades and may therefore aid in revising the generic delimitation in the Nemosenecio-Sinosenecio-Tephroseris assemblage. Although the resolution and support in the ITS phylogeny allow for the transfer of Sinosenecio koreanus and S. newcombei to Tephroseris, the phylogenetic relationships of other Sinosenecio species are currently poorly supported. More detailed studies, involving sequence data from additional DNA regions, are therefore needed before taxonomic changes are made in Sinosenecio. In addition, a larger taxon sampling for Sinosenecio and closely related genera which would include several new species of Sinosenecio that were recently reported (for example, Zhang et al., 2008) should be used to arrive at a stable classification of the Nemosenecio-Sinosenecio-Tephroseris assemblage.
|
||
|
|
|||
|
|
|||
|
WANG et al. ― Recircumscription of subtribe Tephroseridinae
|
441
|
||
|
|
|||
|
Acknowledgements. This study was supported by grants from National Natural Science Foundation of China (30725004), and the Program for New Century Excellent
Talents, Ministry of Education of China (NCET-05-0886).
We thank Drs Wang Yujin and Gao Dahai for their helps in the data analysis and experiments.
LITERATURE CITED
Bain, J.F. and J.L. Golden. 2000. A phylogeny of Packera (Senecioneae; Asteraceae) based on internal transcribed spacer region sequence data and a broad sampling of outgroups. Mol. Phylogenet. Evol. 16: 331-338.
Barkley, T.M. and D.F. Murray. 2006. Tephroseris [M]. In F. O.
N. A. E. Committee (ed.), Flora of North America: north of Mexico Oxford University Press, New York, Oxford, pp. 615-618.
Bremer, K. 1994. Asteraceae: Cladistics and Classification [M], Timber Press, Portland.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
Golden, J.L., YD. Kim, and J.F. Bain. 2001. A re-evaluation of North American Tephroseris and Sinosenecio (Asteraceae: Senecioneae) based on molecular and micromorphological data. Can. J. Bot. 79: 1195-1201.
Greene, E.L. 1897. Senecio newcombei. Pittonia 3: 249.
Huelsenbeck, J.P. and F. Ronquist. 2001. MRBAYES: Bayesian
inference of phylogeny. Bioinformatics 17: 754-755.
Janovec, J.P. and T.M. Barkley. 1996. Sinosenecio mwcombgi (Asteraceae: Senecioneae): A New Combination for a North American Plant in an Asiatic Genus. Novon 6: 265-267.
Jeffrey, C. 1 992. Notes on Compositae, VI: The tribe Senecioneae (Compositae) in the Mascarene Islands with an annotated world check-list of the genera of the tribe. Kew Bull. 47: 49-109.
Jeffrey, C. and Y.L. Chen. 1984. Taxonomic studies on the tribe Senecioneae (Compositae) of Eastern Asia. Kew Bull. 39:
205-446.
Knox, E.B. and J.D. Palmer. 1995. Chloroplast DNA variation and the recent radiation of the giant senecios (Asteraceae) on the tall mountains of eastern Africa. Proc. Nat. Acad. Sci. USA 92: 10349-10353.
Liu, J.Q. 1999. Systematics of the subtribe Tussilagininae of the Senecionae of eastern Asia. Unpublished PhD Dissertation. Beijing: Institute of Botany, Chinese Academy of Sciences.
Liu, J.Q. 2000. Pollen wall ultrastructures of the subtribe Tussilagininae (Asteraceae: Senecioneae) of eastern Asia and their systematic and taxonomic significance. J. Wuhan Bot. Res. 18: 461-465.
Liu, J. Q. 2001 . Floral microcharacters of the subtribe Tussilagininae (Asteraceae: Senecioneae) of eastern Asia and their systematic and taxonomic significance. Bull. Bot.
Res. 21: 58-67.
Liu, J. Q. 2004. Uniformity of karyotypes in Ligularia
|
(Asteraceae: Senecioneae), a highly diversified genus of the eastern Qinghai-Tibet Plateau highlands and adjacent areas.
Bot. J. Linn. Soc. 144: 329-342.
Liu, J.Q., T.G. Gao, Z.D. Chen, and A.M. Lu. 2002. Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae). Mol. Phylogenet. Evol.
23: 307-325.
Liu, J.Q., Y.J. Wang, A.L. Wang, H. Ohba, and R.J. Abbott.
2006. Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol.
Phylogenet. Evol. 38: 31-49.
Nordenstam, B. 1 978. Taxonomic studies in the tribe Senecioneae (Compositae). Opera Botanica 44: 1-83. Lund.
Nordenstam, B. 2007. Tribe Senecioneae. In J.W. Kadereit and
C. Jeffrey (eds.), The Families and Genera of Vascular
Plants Vol. VIII, pp. 208-241.
Panero, J.L., J. Francisco-Ortega, R.K. Jansen, and A. Santos-Guerra. 1999. Molecular Evidence for Multiple Origins of Woodiness and a New World Biogeographic Connection of the Macaronesian Island Endemic Pericallis (Asteraceae:
Senecioneae). Proc. Nat. Acad. Sci. USA 96: 13886-13891.
Pelser, P.B., B. Gravendeel, and van der R. Meijden. 2002. Tackling speciose genera: species composition and phylogenetic position of Senecio sect. Jacobaea (Asteraceae) based on plastid and nrDNA sequences. Am. J.
Bot. 89: 929-939.
Pelser, P.B., B. Gravendeel, and R. van der Meijden. 2003. Phylogeny reconstruction in the gap between too little and too much divergence: the closest relatives of Senecio jacobaea (Asteraceae) according to DNA sequences and
AFLPs. Mol. Phylogenet. Evol. 29: 613-628.
Pelser, P.B., B. Nordenstam, J.W. Kadereit, and L.E. Watson.
2007. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon 56: 1077-1104.
Posada, D. and K.A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817-818.
Ronquist, F. and J.P. Huelsenbeck. 2003. MRBAYES 3:
Bayesian phylogenetic inference under mixed models.
Bioinformatics 19: 1572-1574.
Swenson, U. and K. Bremer. 1997. Patterns of Floral Evolution of Four Asteraceae Genera (Senecioneae, Blennospermatinae) and the Origin of White Flowers in
New Zealand. Syst. Biol. 46: 407-425. Swofford, D.L. 2002. PAUP*: Phylogenetic Analyses Using
Parsimony (* and Other Methods), Version 4 [M]. Sinauer & Associates, Sunderland, Massachusetts.
Taylor, R.L. and G.A. Mulligan. 1968. Flora of the Queen Charlotte Islands, 2. Cytological aspects of the vascular plants. Res. Branch, Canad. Dept. Agric., Monogr. 4 part 2.
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and
D. G. Higgins. 1997. The Clustal-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analyses tools. Nucleic Acids Res. 24: 4876-4882.
|
||
|
|
|||
|
|
|||
|
442
|
Botanical Studies, Vol. 50, 2009
|
||
|
|
|||
|
Wagstaff, S.J. and I. Breitwieser. 2004. Phylogeny and Classification of Brachyglottis (Senecioneae, Asteraceae): An Example of a Rapid Species Radiation in New Zealand.
Syst. Bot. 29: 1003-1010.
|
White, T.J., T. Bruns, S. Lee, and J.W. Taylor. 1990.
Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics [M]. In M. Innis, D. Gelfand, J. Sninsky, and T. White (eds.), PCR Protocols. Academic Press, San Diego, CA., pp. 315-322.
|
||
|
Wagstaff, S.J., I. Breitwieser, and U. Swenson. 2006. Origin and relationships of the austral genus Abrotanella (Asteraceae) inferred from DNA sequences. Taxon 55: 95-106.
|
|||
|
Zhang, D.G., Y. Liu, and Q.E. Yang. 2008. Sinosenecio jishouensis (Compositae), a new species from north-west
Hunan, China. Bot. Stud. 49: 287-294.
|
|||
|
Weber, W.A. and A. Love. 1981. New combinations in the genus Packera. Phytologia 49: 44-50.
|
|||
|
|
|||
|
狗舌草亞族複合群(菊科:千里光族)的屬間界限與ITS
分子證據的衝突
王留陽1,2,5 Pieter B. PELSER3 Bertil NORDENSTAM4 劉建全2
|
|||
|
|
|||
|
1中國科學院西北高原生物研究所進化與適應實驗室
2中國蘭州大學生命科學院乾旱與草地生態重點實驗室 3 Oklahoma State University, Botany Department, 104 Life Sciences East Stillwater, Oklahoma 74078-3013, USA
4 Swedish Museum of Natural History, P. O. Box 50007, SE-104 05 Stockholm, Sweden
5北京中國科學院研究生院
|
|||
|
|
|||
|
傳統菊科千里光族狗舌草亞族主要包括狗舌草屬、華千里光屬和羽葉千里光屬三屬,這裏我們暫
稱?“狗舌草亞族複合群”。目前這三個屬的親緣關係和系統位置存在較大分歧,需要進一步修訂。 在本研究中,我們新報導了 13個種的19條核糖體內轉錄間隔區ITS序列;並結合Genbank已報導序 列,對該複合群27個種(覆蓋了這三個屬所有的組和亞組)和款東亞族內近緣屬的7個代表種的核糖 體內轉錄間隔區ITS構建了分子系統發育樹。研究發現華千里光屬和狗舌草屬均非單系起源。華千里光 屬四個種,包括該屬的模式種(S. eriopodus, S. hederifolius, S. homogyniphyllus禾口 S. subcoriaceus)與橐 吾屬、垂頭菊屬和蟹甲草屬等近緣屬的代表種聚為一支,即分支B ,但支援率不高;而該屬其他10個 代表種則與狗舌草屬的13個種和羽葉千里光屬的3個種共同組成分支A ,並得到較高的自展支持。主 要分支A含有兩個穩定的亞分支C和D :其中亞分支C包括狗舌草屬的10個種與華千里光屬的兩個 種S. newcombei和S. koreanus '而亞分支D包括了所有三個屬的其他11個種。另外'亞分支D內, T. changii與華千里光屬的4個代表種聚為一支,而羽葉千里光屬的3個代表種則單獨組成一個單系分 支。結合先前的有關外部宏觀和微觀性狀特徵的研究與本研究中的ITS分子證據,這三個屬的屬間界限 需要進行適當調整:newcombei和S. koreanus兩個種應放在狗舌草屬內;而華千里光屬也應包括本? 究中分支B內一些近緣屬的代表種;本?究所涉及到華千里光屬和T. changii應歸併至羽葉千里光屬, 或成立一個新屬。但無論如何,這三個屬間界限仍需要更多形態學和分子證據,需要進一步的調查和研 究。 |
|||
|
|
|||
|
關鍵詞:菊科;款東亞族;華千里光屬;狗舌草屬;羽葉千里光屬;分子系統發育;核糖ITS區域。
|
|||
|
|
|||