478
Botanical Studies, Vol. 50, 2009
outgroups. Leaf materials were preserved in silica gel bags immediately after sampling. Voucher specimens were deposited at the herbaria of TAI or HAST.
DNA extraction, PCR and sequencing
Total genomic DNA was extracted from dried leaf tissue, following the protocol of Vijayan and Tsou (2008). Primers used for PCR amplification and sequencing of
the RPB2 introns 12-16 and 23 are listed in Table 2. PCR
was performed in a 50 fiL final volume containing 100 ng DNA, 0.2-0.6 foM of both primers, 200 fiM of each dNTP, 0.5 U Taq polymerase and 1X buffer (Viogene, Inc.). The PCR cycle was with an initial 5 min denaturation at 95oC, followed by 35 cycles of 1 min denaturation at 95oC, 1 min 20 sec annealing at 58oC to 63oC, 1 min 20 sec extension at 72oC, and with a final extension of 7 min at 72 。C. PCR products were purified using QIAquick® PCR Purification Kit (Qiagen Gmbh, Inc.). Automatic sequencing was conducted with an ABI PRISM® 3700 DNA Sequencer.
Data analysis
Sequences were assembled from both directions. Nucleotide sequences of introns 12-16 and 23 were aligned with CLUSTAL_X v1.83 (Thompson et al., 1997). The phylogenetic analyses were performed with the maximum parsimony (MP) and neighbor joining (NJ) methods using PAUP v0.4b10 (Swofford, 2001). In the MP analysis, the most strict consensus tree was obtained by performing a branch-and-bound searching. Indels were treated as single base changes and gaps were considered as the fifth base. In the NJ analysis, distances were measured with an 'uncorrected ("p")' index. Indels were also treated as a single base change but gaps were considered as missing. Branch supports of both phylogenetic trees were obtained by 1000 replicates of bootstrapping. Additionally, the genetic distances between taxa were calculated with the 'uncorrected ("p")' index under the distance criterion of
PAUP.
RESULTS
Introns 12-16 of RPB2 of samples examined were 954 bp long. No indels were detected. The intron 23 was 992 to 1001 bp long in all the samples except for two samples of C. sinensis var. assamica, i.e., A-Thai-2 and A-Ind-3, in which a long insertion of 284 bp was found. In the analysis, the long insertion was treated as a single mutation and a data matrix of 1923 bp for 35 OTUs was obtained.
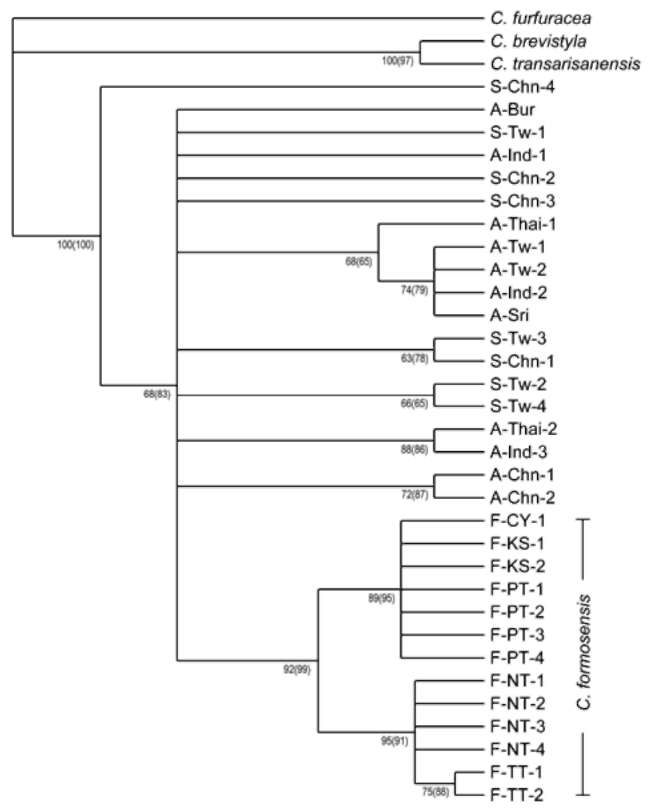
Of 1923 bp sequences, 166 bp were variable and 75 bp were parsimoniously informative. Branch-and-bound search found 1896 equally parsimonious trees. The most strict consensus tree was identified with 233 steps. A high consistency index (CI) value 0.92 and low homoplasy index (HI) value 0.25 indicated low levels of homoplasy. MP and NJ trees were the same in topology though the branch supports might be different (Figure 2). All 13 samples of the Taiwanese wild tea formed a distinct
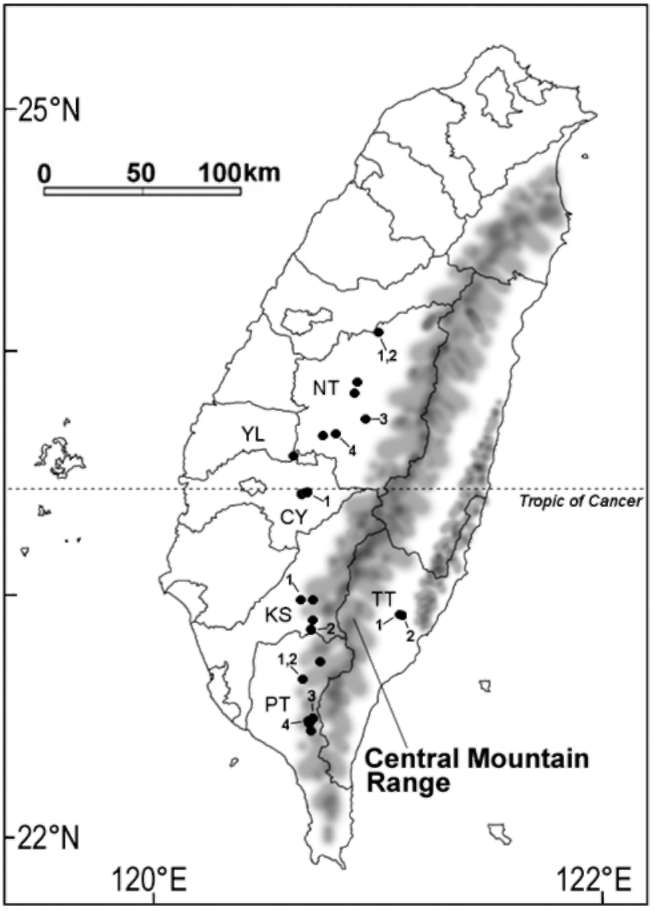
Figure 1. Distribution and sampling sites of Camellia formosensis. Solid circles: distribution sites based on specimens of TAI, TAIF, HAST and TNM herbaria. DNA sampling sites are marked with the running number. NT, Nantou County; YL, Yunlin County; CY, Chiayi County; KS, Kaohsiung County; PT, Pingtung County; TT, Taitung County.
provided much insight into the phylogeny of Camellia in an earlier studies (Xiao, 2001; Xiao and Parks, 2003); however, no materials from the Taiwanese wild tea were included. We thus reconstructed the phylogeny of the Taiwanese wild tea and its allies by using the same DNA segments, the RPB2 introns 12-16 and 23, and with a much more extensive sampling. The results are here documented.
MATERIALS AND METHODS
Samples
Thirty-five samples were included in the study, including 13 samples of the Taiwanese wild tea, eight samples of C. sinensis var. sinensis and 11 samples of C. sinensis var. assamica, all belonging to the section Thea (Table 1). Among the 13 samples of the Taiwanese wild tea, 11 were collected from wild populations (Figure 1) while the other two were obtained from the tea germplasm garden (Table 1). Three species―C. furfuracea (sect. Furfuracea), C. transarisanensis (sect. Theopsis), and C. brevistyla (sect. Paracamellia)——were chosen as