CHEN and SUN ― Arabidopsis atToc33 and atToc34 genes
149
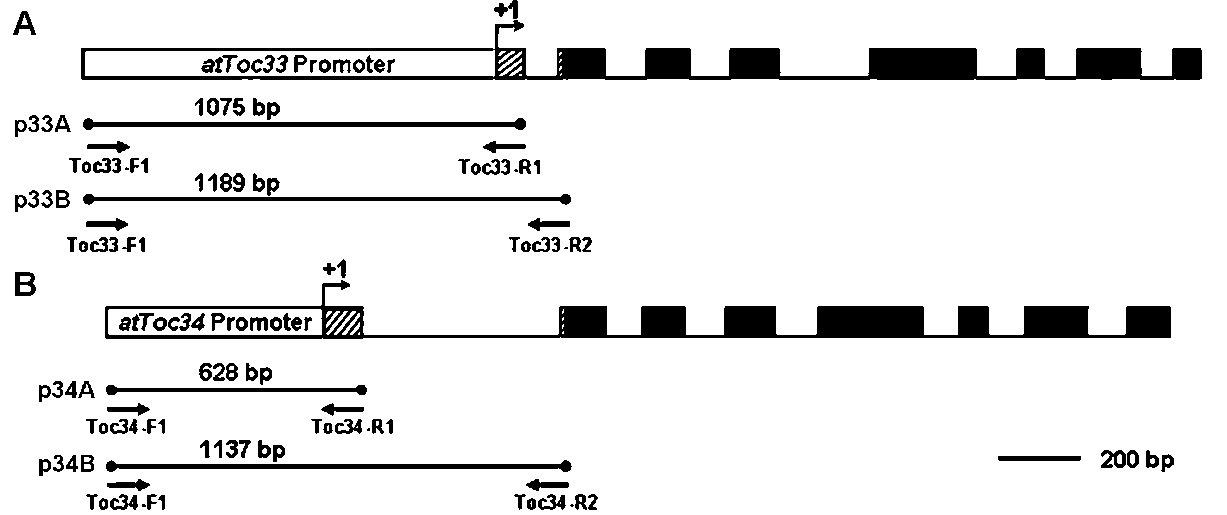
Figure 1. Gene structure and upstream construct of atToc33 and atToc34. The promoter (-1020 to -1), 5' UTR (+1 to +55 and +158 to +169), and leader intron (+56 to +157) sequences of atToc33 gene were shown in panel A. The promoter (-532 to -1), 5' UTR (+1 to +96 and +590 to +605), and leader intron (+97 to +589) sequences of atToc34 gene were shown in panel B. The p33A and p34A constructs consist of the promoter and 5' UTR sequences prior the lead intron of atToc33 and atToc34, respectively. The p33B and p34B constructs contain additional leader intron sequences of atToc33 and atToc34, respectively. Arrows indicated the locations and names of primers. Open box, promoter sequence. Hatched box, 5' UTR sequence. Black box, coding sequence. Bold underline, intron sequence. +1, transcription start site.

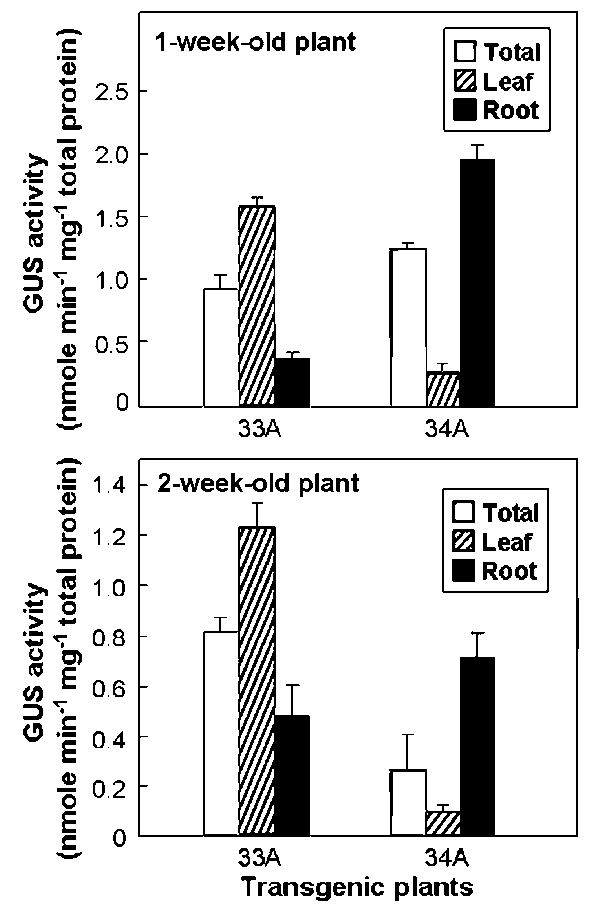
and so did 34A-5-1, 34A-6-2, 34A-8-5, 34A-10-2, and 34A-12-5 plants (data not shown). Therefore, 33A-6-2 and 34A-5-1 plants were used as representatives in Figure 3. The atToc33 expression in leaves was approximately three times higher than in roots in both 1-week-old and 2-week-old 33A-6-2 plant. However, the overall atToc33 expression in whole plants (the open boxes of Figure 3) of both ages was similar. This revealed that the constant expression level of atToc33 observed in Figure 2 was a compromised result obtained from mixed plant tissues. In fact, atToc33 was preferentially expressed in leaves and did not change significantly in roots. By contrast, atToc34 expression in roots was about six times higher than in leaves in both 1-week-old and 2-week-old 34A-5-1 plants. Agreeing with the results in Figure 2, the overall atToc34 expression yield in 1-week-old 34A-5-1 plants was approximately 3 to 4 times higher than 2-week-old plants. These results indicate that atToc34 is preferentially expressed in roots and young plants. In other words, atToc34 is regulated in both age- and tissue-specific manners.
Intron effect in transgenic plants expressing atToc33 promoter- and atToc34 promoter-GUS fusion genes
Leader intron (also called proximal intron) has been shown to have some influence in determining the level and pattern of gene expression in Arabidopsis (Norris et al., 1993; Jeong et al., 2006). The mechanism of intron-mediated enhancement (IME) is largely unknown, however, it was hypothesized that IME might increase transcription efficiency or mRNA stability (Rose et al., 2008). As atToc33 and atToc34 have similar gene
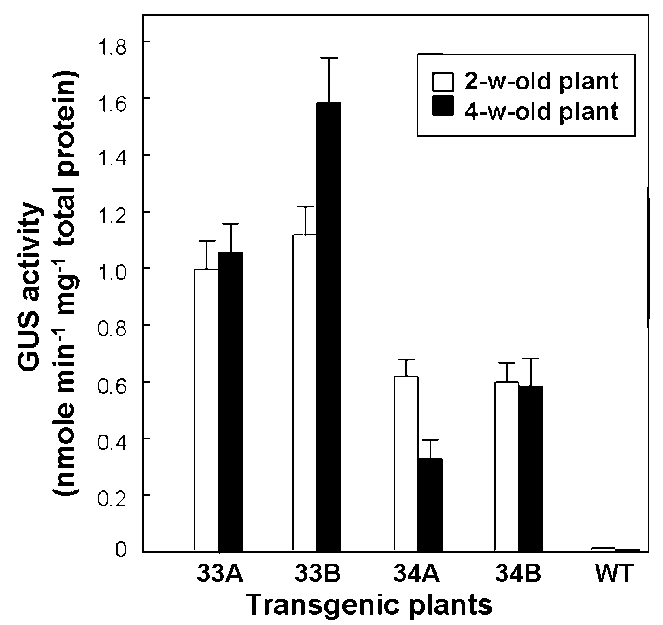
Figure 2. Expression of atToc33 and atToc34 genes in different developmental stages. The GUS activity of 1- to 4-week-old transgenic plants 33A and 33B was determined. Data are means 士SD of three repeat experiments from at least four individual transgenic lines (33A-2-6, 33A-4-5, 33A-6-2, 33A-7-3, 34A-5-1, 34A-6-2, 34A-8-5, 34A-10-2, and 34A-12-5). P values < 0.001 (Tukey test) were considered to represent significant differences.
contrast, atToc34 expression was related to plant age, with highest promoter activity in the young seedling (1-week國 old) and a compromised yield in the mixed tissues of mature plants (2- to 4-week-old). This indicates that atToc34 is modulated in an age-dependent manner.
Expression of atToc33 and atToc34 genes in vegetative tissues
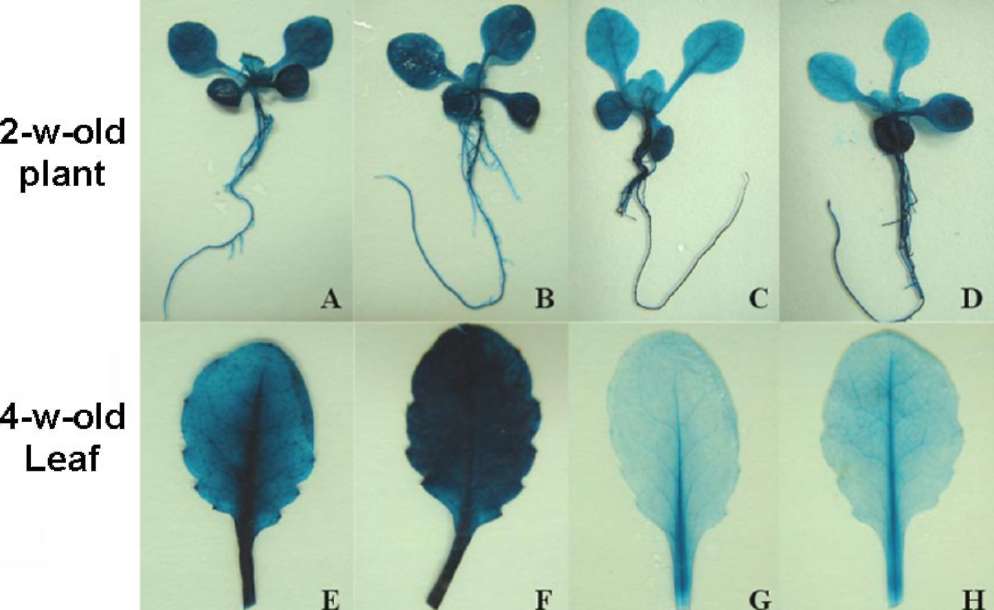
Next, we asked if atToc33 and atToc34 are differentially expressed in vegetative tissues. The GUS activity in leaves and roots of four 33A lines and five 34A lines was compared. The 33A-2-6, 33A-4-5, 33A-6-2, and 33A-7-3 plants revealed similar expression pattern by fluorometric quantification and histochemical staining of GUS activity,