SHIH et al. — Tissue- and cellular localization
185
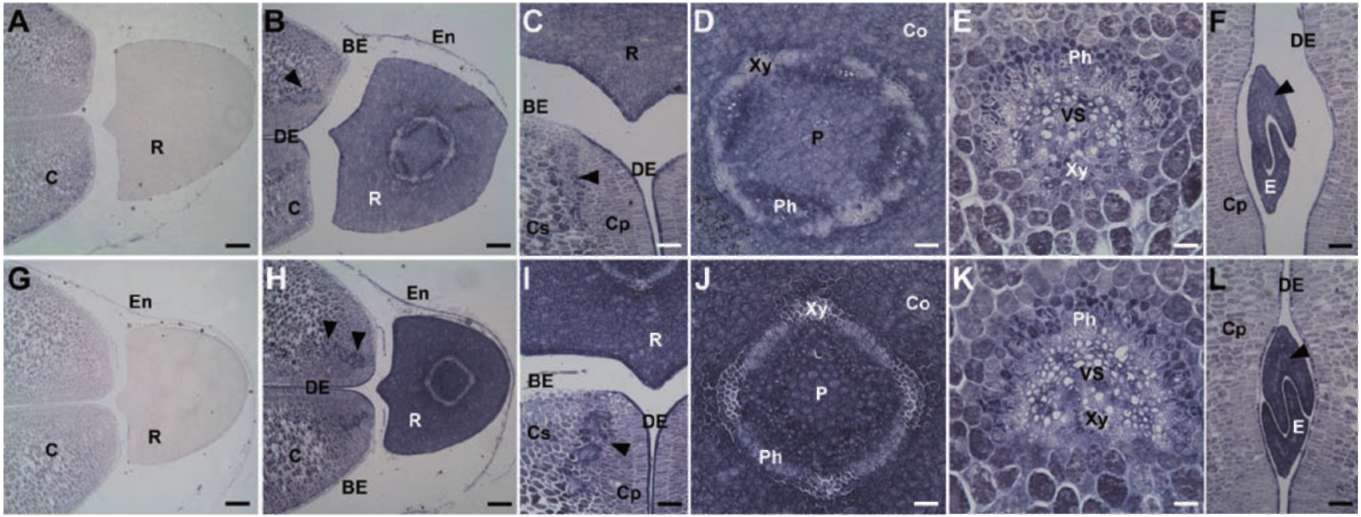
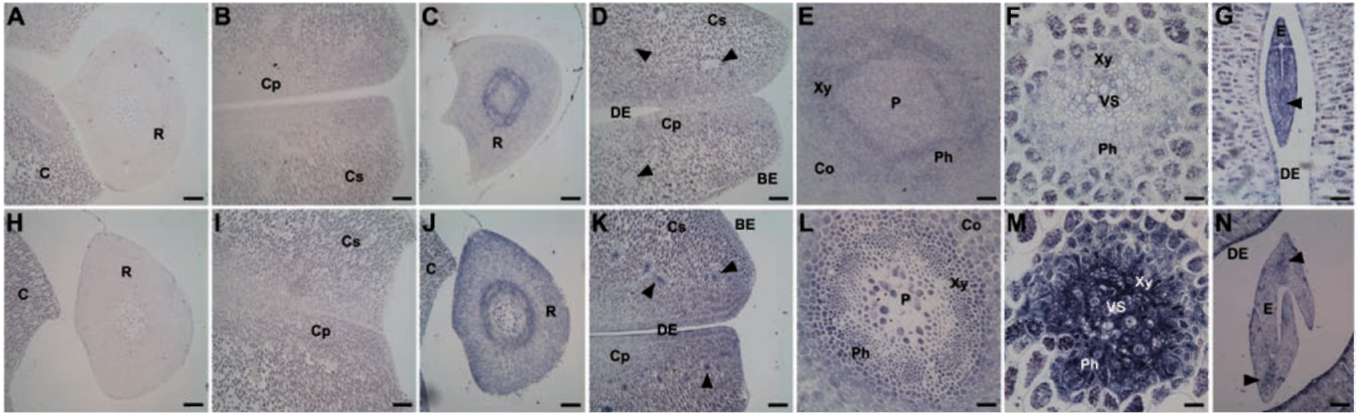
predicted molecular mass is 9 kDa, at pI 7.5. The spatial expression patterns of GmPM11 were analyzed in fresh or 4-day PD seeds at 35 DAF (Figure 1). In fresh seeds, GmPM11 transcripts highly accumulate in vascular tissues of radicle but are barely detected in cotyledon vascular strands and radicle cortex (Figure 1B to E). In 4-day PD seeds, GmPM11 mRNAs accumulate in all seed tissue, including the cotyledon and embryo axis (Figure 1F to J). Strong signals are observed in vascular tissues and in the adaxial epidermis cells of cotyledon.
Several Em6-type LEA genes had been studied by in situ hybridization. Carrot Emb-1 mRNA was preferentially detected in the procambial meristematic region of somatic or zygotic embryos from the globular to mature torpedo stages and was greatly accumulated in cotyledons at a later stage (Wurtele et al. , 1 993 ). Sunflower ds11 transcripts appeared to accumulate to high levels in early embryogenesis. In mature seeds, the distribution of ds11 mRNA showed marked localization in vascular tissues of the procambium or cotyledon (Prieto-Dapena et al., 1999). Arabidopsis AtEm6 transcripts were detected throughout the entire embryo in provascular tissues and shoot apical meristem (Vicient et al., 2000). Therefore, the in situ hybridization studies of other Em6-type LEA I genes showed a similar signal distribution in seeds or embryos similar to what we found. These results suggested that the messages of Em6-type LEA I genes appear from the early to middle stages of embryogenesis and reach a plateau at the late developmental stage, with preferential accumulation in vascular tissues of the procambium of the cotyledon or embryo axis.
Localization of Group II LEA messages
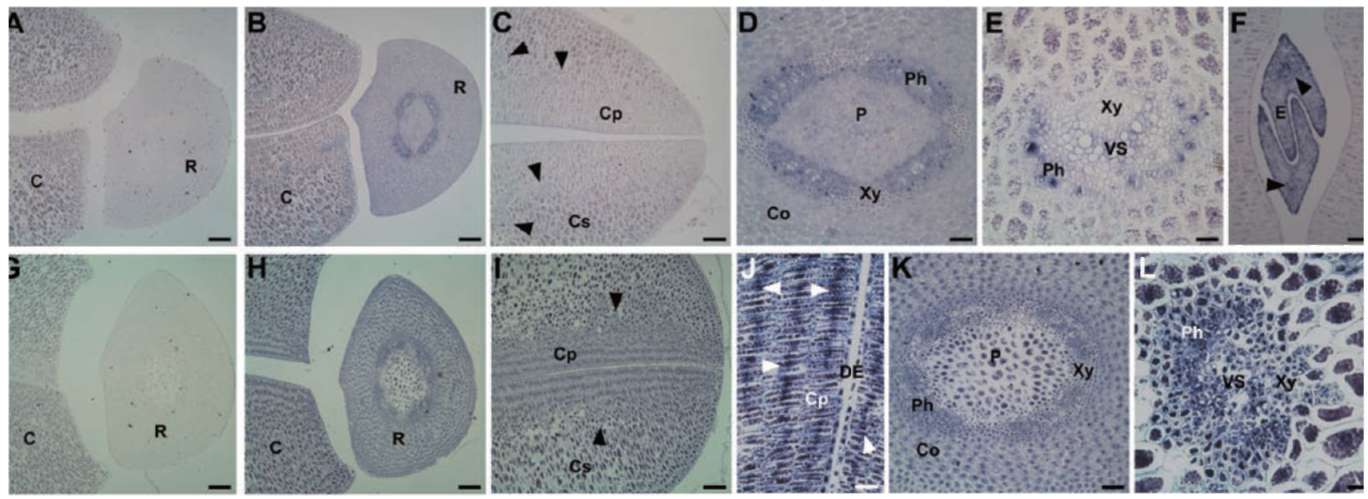
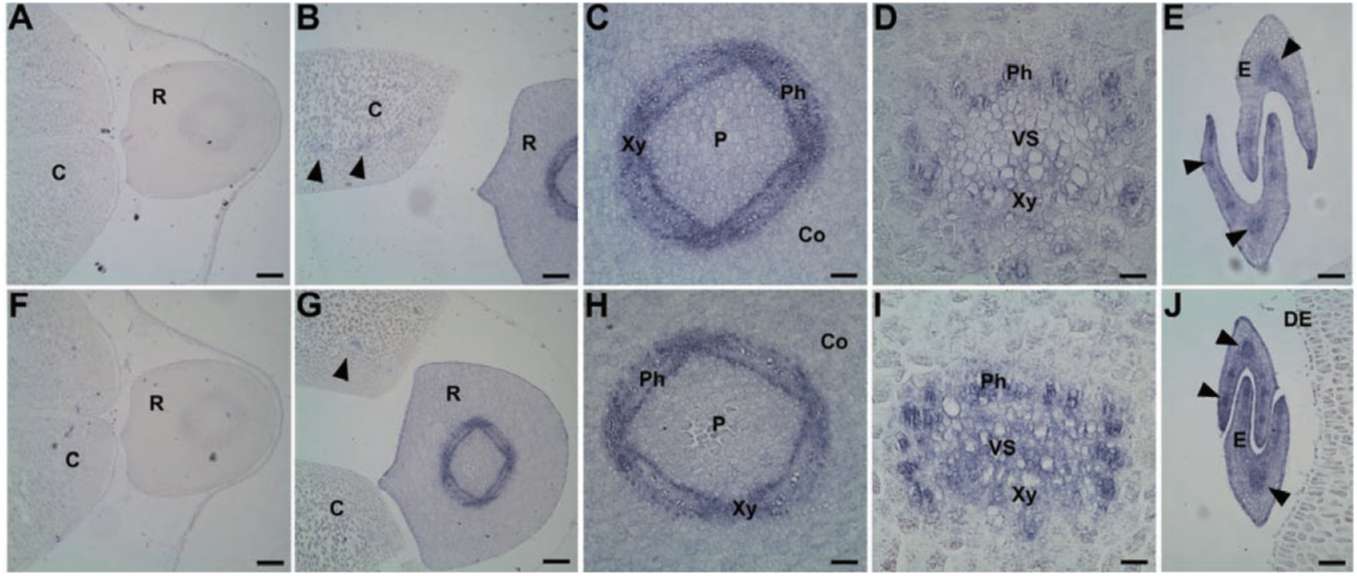
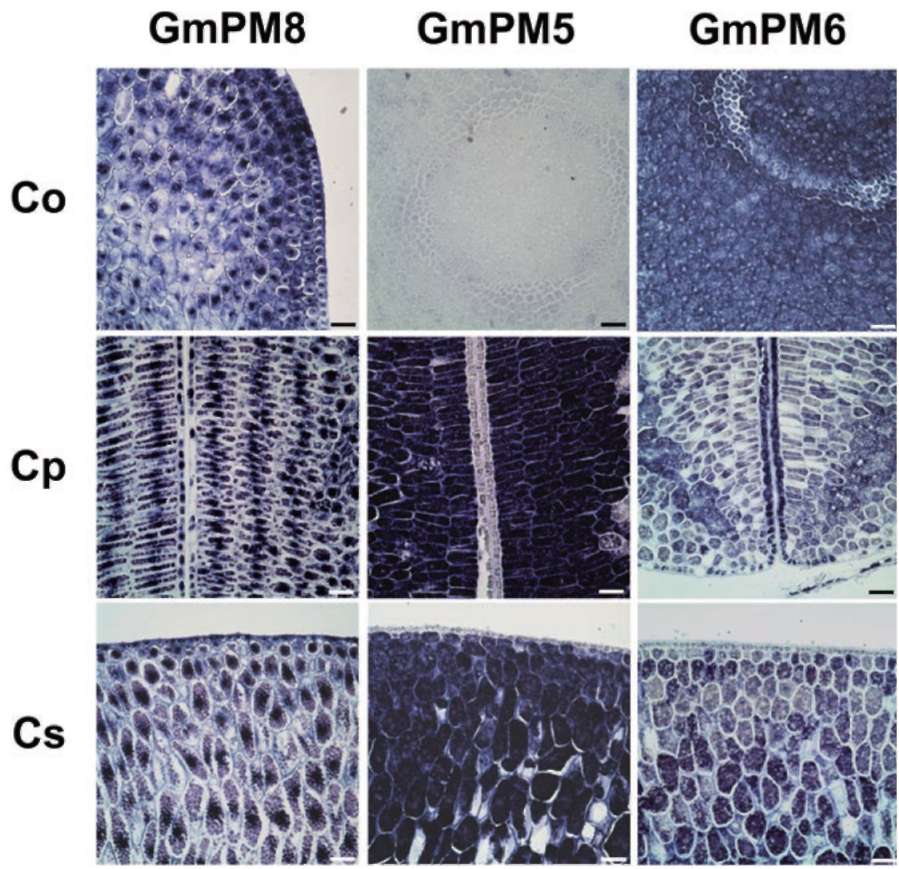
Group II LEA proteins have 3 specific, characteristic sequence "modules" (i.e., K-domain, Y-domain, and S-segment) that define this class of protein (Close, 1997). In the present study, we used these soybean dehydrin genes (i.e., GmPM6/7 and GmPM12) to illustrate their spatial patterns. GmPM6/7 (accession nos. M9401 2 and L00921) share 96% cDNA sequence identity. The deduced protein sequences of GmPM6/7 belong to Y2K-type dehydrins, with molecular mass of 31/32 kDa and pI 6.6/6.5, respectively. The mRNA of GmPM6/7 (the
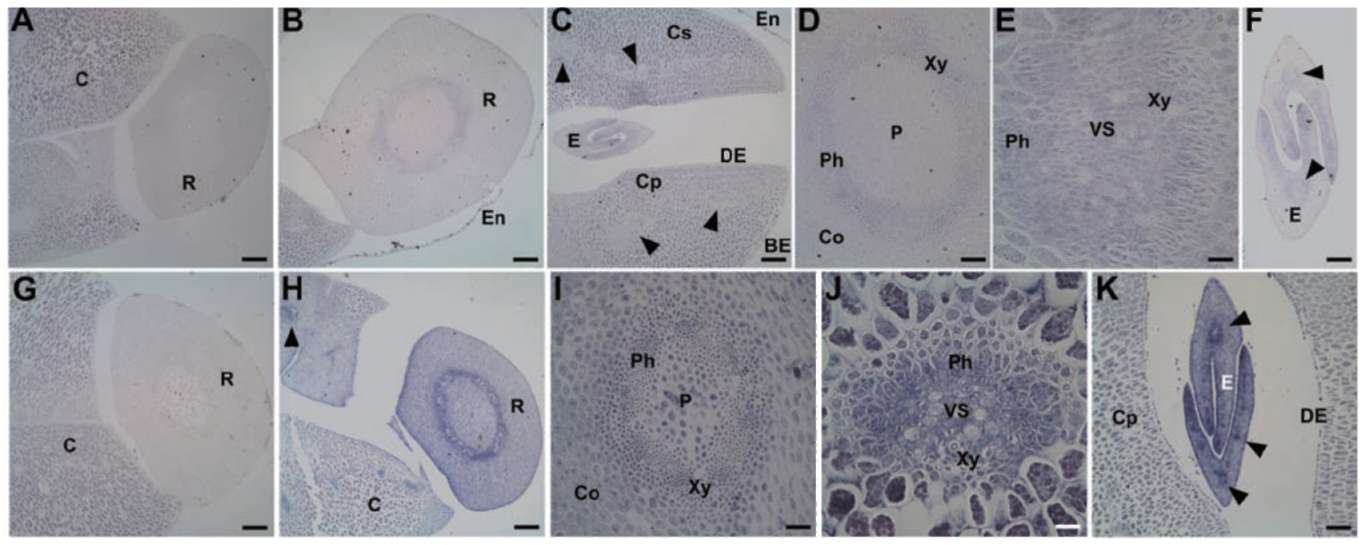
2 transcripts cannot be distinguished because of high identity) accumulates in vascular strands, mesophyll around vascular tissues, and adaxial epidermis of cotyledon, epicotyl, as well as cortex, pith, and phloem cells of radicle in fresh seed (Figure 2B to F). By contrast, no or few GmPM6/7 transcript is detected in the abaxial epidermis of cotyledon (Figure 2B and C) and radicle pericycle (Figure 2D). The mRNA expression of GmPM6/7 mRNA is greatly increased in the same regions in PD seeds as in fresh seeds (Figure 2G to L). The mRNA level of GmPM6/7 is increased slightly in radicle pericycle and of cotyledon vascular strands after drying (Figure 2J and K).
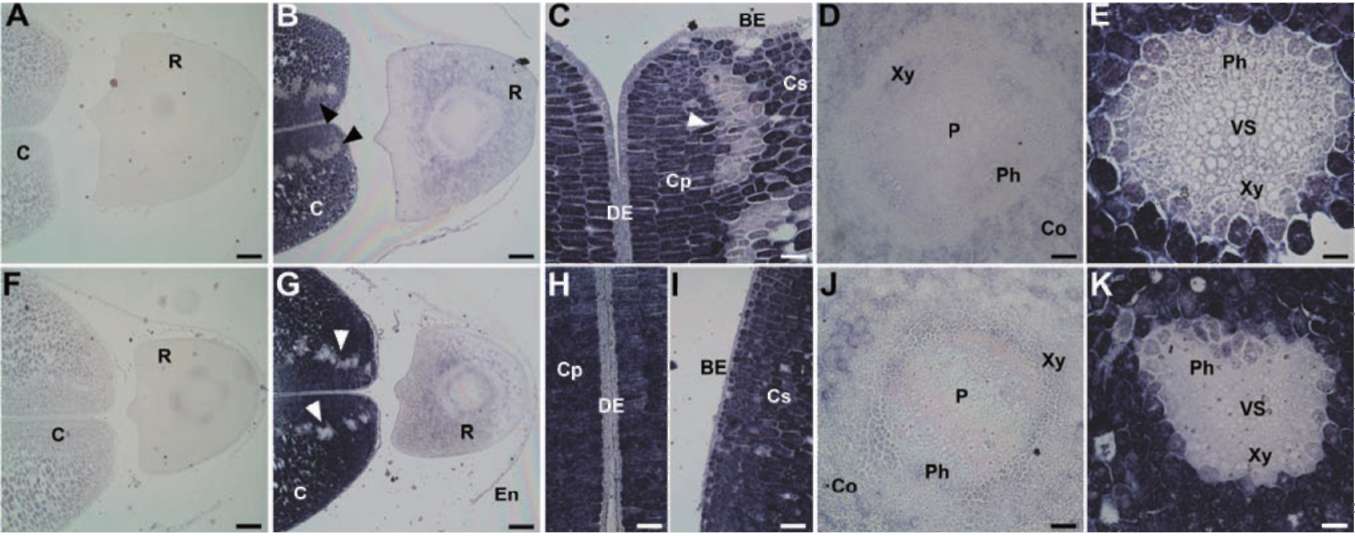
The deduced protein sequence of GmPM12 (accession
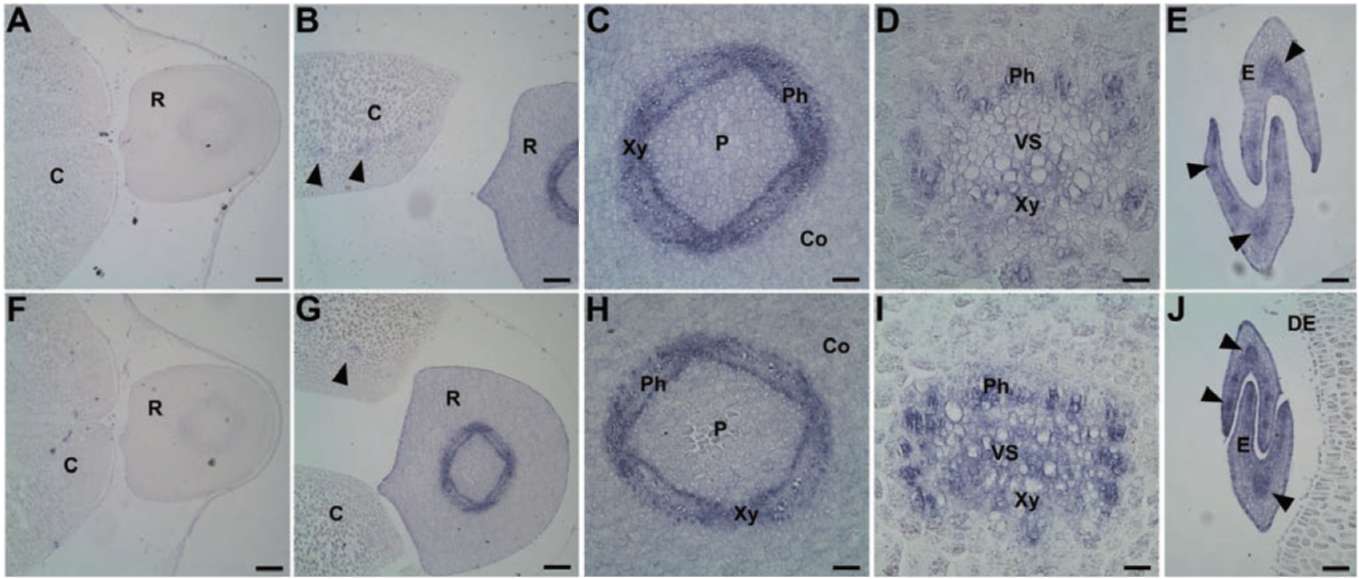
no. AF004807) shows it to be a Y2SK2-type dehydrin. The molecular mass is 10 kDa at pI 10. In fresh seeds, no or few transcript of GmPM12 in seeds (Figure 3B-F). In PD seeds, the expression of GmPM12 is significantly increased in vascular tissues of the cotyledon and embryo axis (Figure 3H to K).
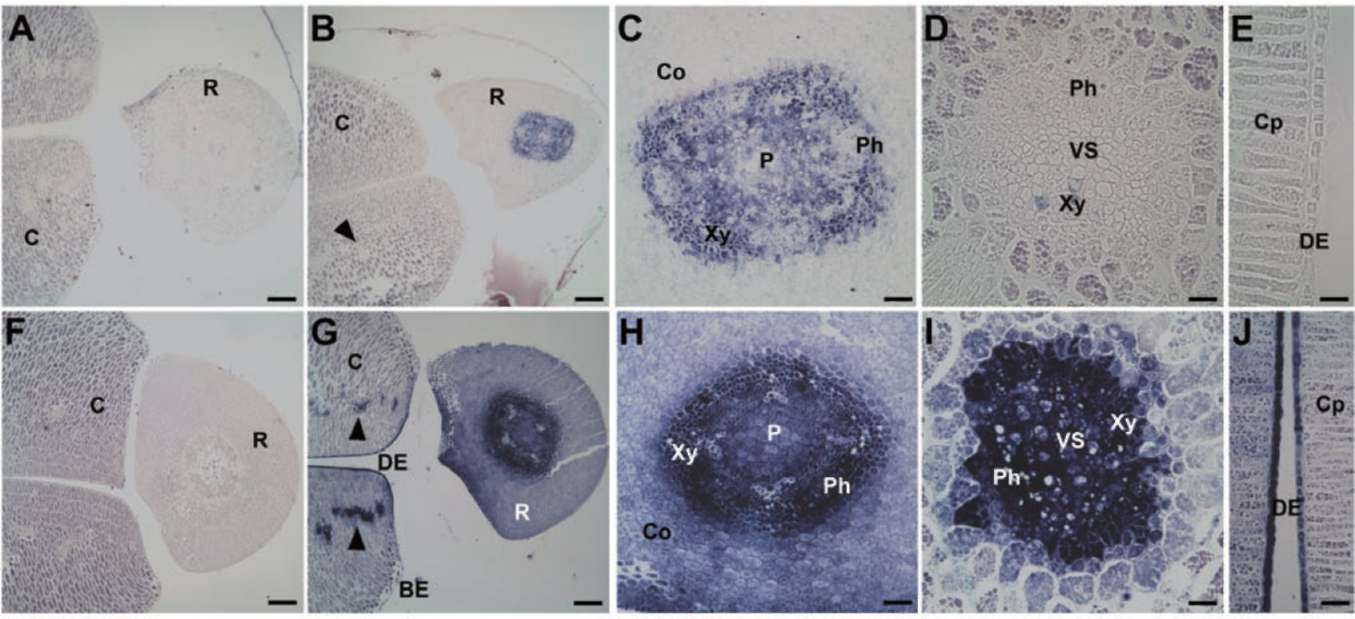
Figure 1. Localization of soybean GmPM11 transcripts in fresh (panel A to E) or 4-day pod-dried (PD) (panel F to J) soybean seeds. In situ hybridization was carried out with transverse sections of seeds in this and following figures. A sense probe was used in panel A and F and an antisense probe in the remaining panels. Bar=200 (im for panel A, B, F, and G, 60 (im for panel C and H, 40 (im for panel E and J, and 20 (im for panel D, and I. Abbreviations (all the abbreviations is noted at first time): BE, abaxial epidermis; C, cotyledon; Co, cortex; Cp, palisade tissue of cotyledon; DE, adaxial epidermis; P, pith; Ph, phloem; R, radicle; VS, vascular strand; Xy, xylem. Arrowheads indicate cotyledon vascular strands.