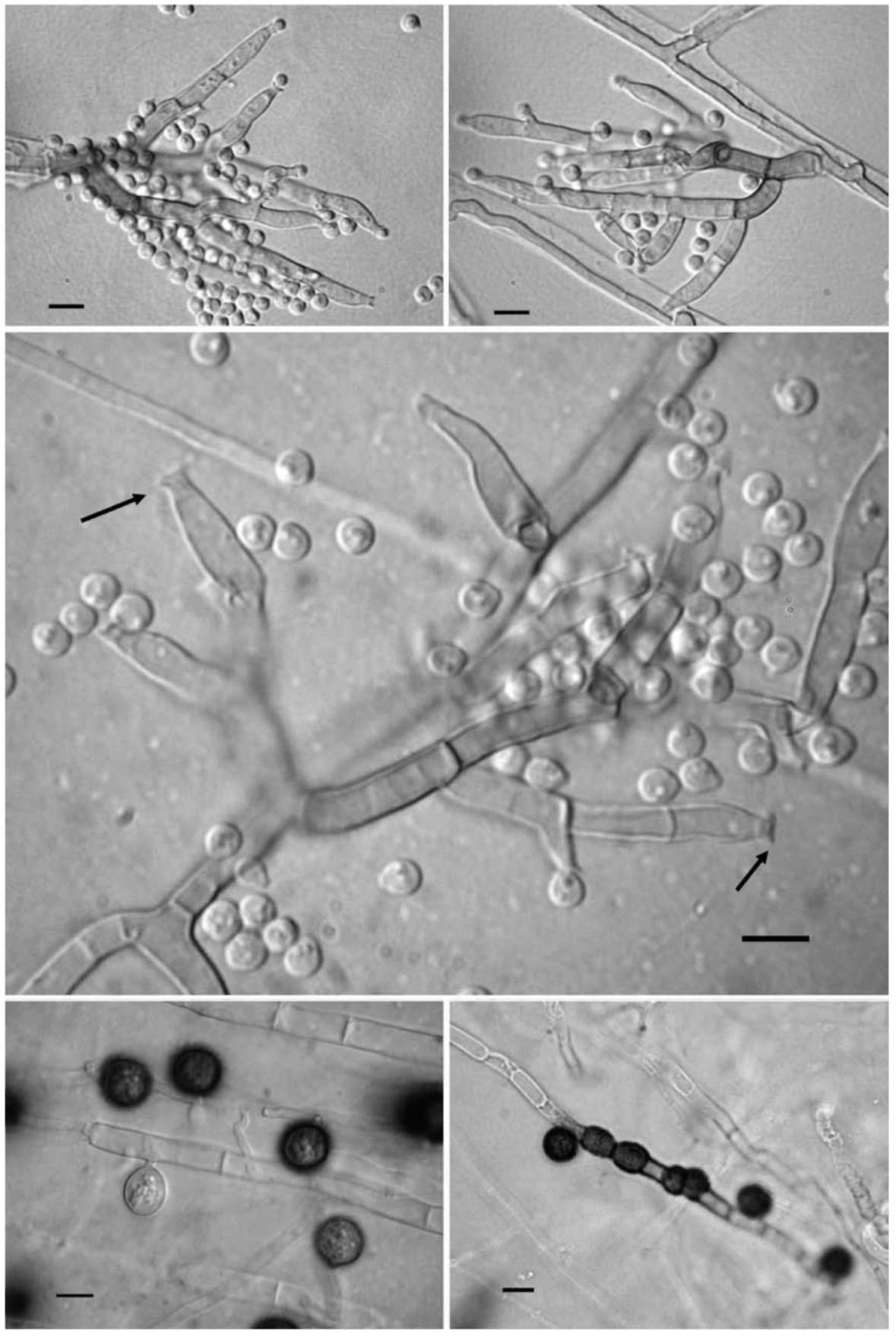
Gams, W. and V. Holubova-Jechova. 1976. Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Stud. Mycol. 13: 59-72.
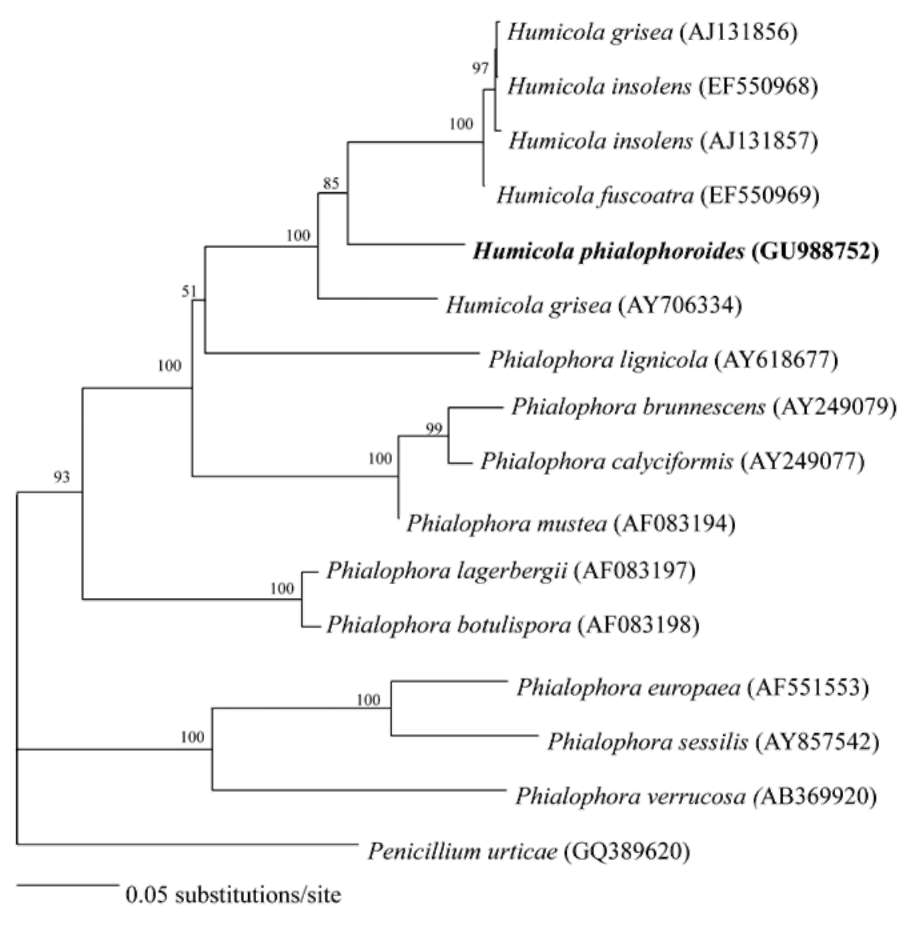
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120.
Ko, W.H. 1981. Reversible change of mating type in Phytoph-thora parasitica. J. Gen. Microbiol. 125: 451-454.
Ko, W.H., Y.J. Tsou, Y.M. Ju, H.M. Hsieh, and P.J. Ann.
2010a. Production of a fungistatic substance by Pseudallescheria boydii isolated from soil amended with vegetable tissues and its significance. Mycopathologia 169: 125-131.
Ko, W.H., Y.J. Tsou, M.J. Lin, and L.L. Chern. 2010b. Activity
and characterization of secondary metabolites produced by a new microorganism for control of plant disease. New Bio-technol. 27 (in press).
Nicoli, R.M. and A. Russo. 1974. The genus Humicola Traaen and the closely related genera (Hyphomycetes). Nova Hedw 3-4: 737-797.
Rambaut, A. 1996. Se-Al Version 1.0 Alpha 1. Sequence Alignment Editor. Oxford, UK: Department of Zoology, University of Oxford.
Saitou, N. and M. Nei. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol.
Biol. Evol. 4: 406-425.
Schol-Schwarz, M.B. 1970. Revision of the genus Phialophora (Moniliales). Persoonia 6: 59-94.
Swofford, D.L. 1998. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (*and Other Method). Sunderland, Massachusetts: Sinauer Associates.
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and D.G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882.
Wang, P.H., Y.S. Chen, M.J. Lin, Y.J. Tsou, and W.H. Ko. 2010. Severe decline of wax apple trees caused by Fusari-um solani in northern Taiwan. Bot. Stud. 51: 75-80.
White, T.J., T. Bruns, S. Lee, and J.W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (eds.), PCR Protocols: A Guide to
Methods and Applications. Academic Piers, San Diego, pp.
315-322.