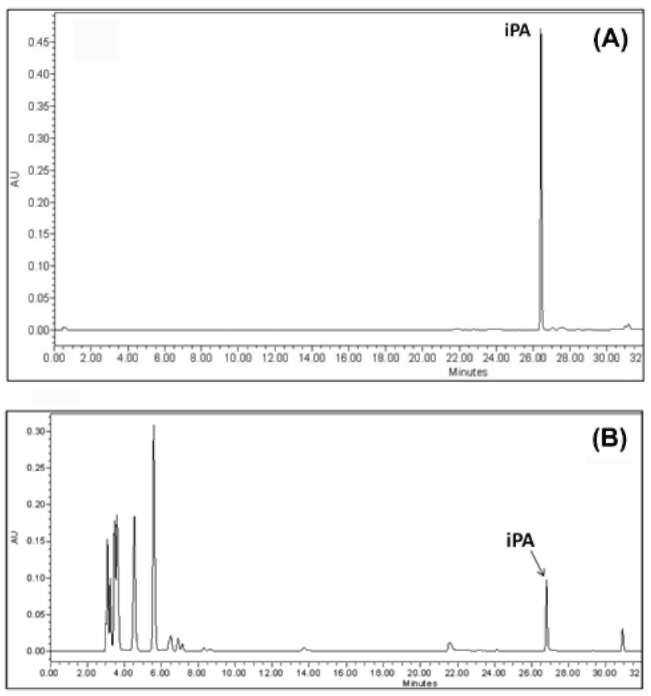
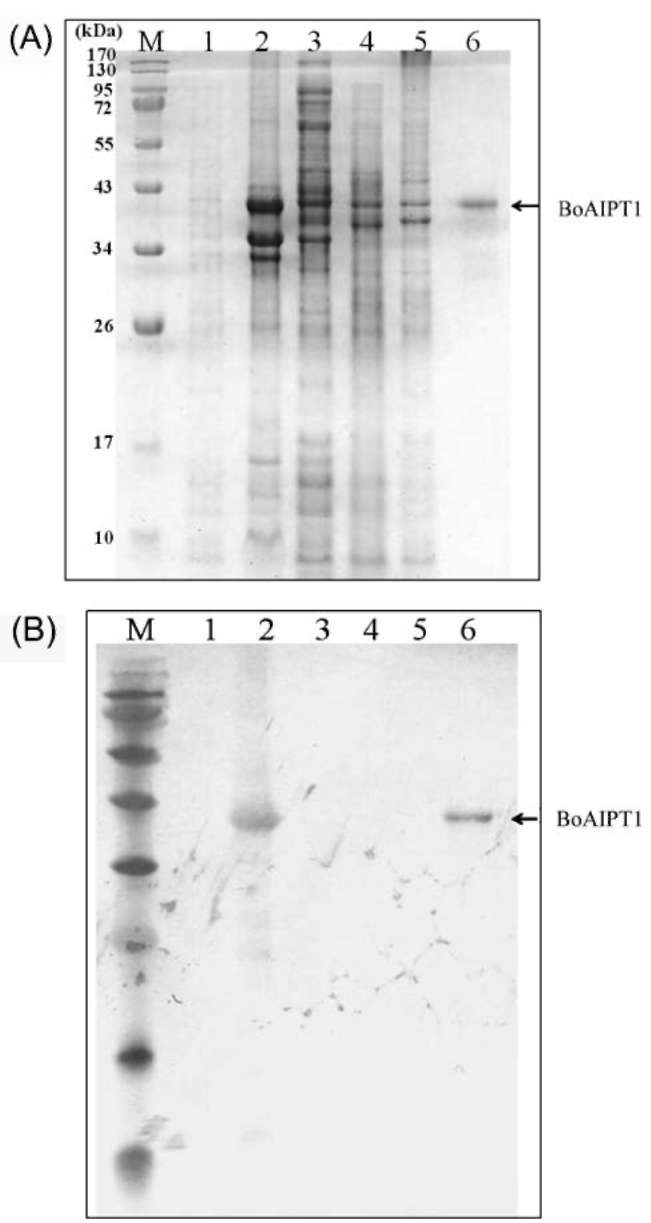
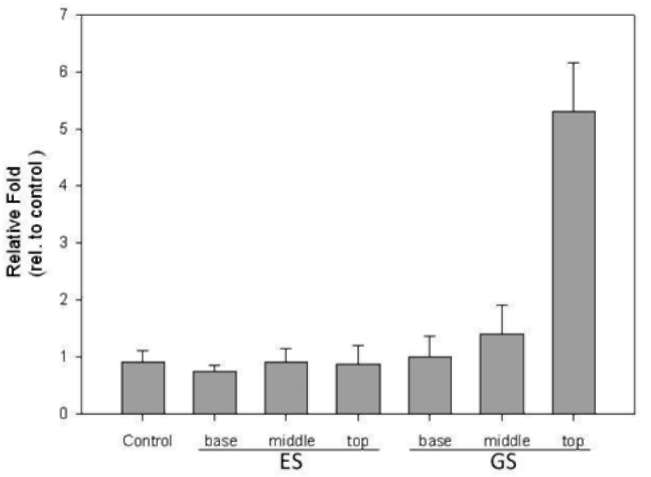
|
|
Chen, L.F.O., J.Y. Hwang, Y.Y. Charng, C.W. Sun, and S.F.
Yang. 2001. Transformation of broccoli (Brassica oleracea var. italica) with isopentenyltransferase gene via Agrobac-terium tumefaciens for postharvest yellowing retardation. Mol. Breed. 7: 243-257.
Chu, H.M., T.P. Ko, and A.H. Wang. 2009. Crystal
structure and substrate specificity of plant adenylate isopentenyltrans-ferase from Humulus lupulus: distinctive binding affinity for purine and pyrimidine nucleotides. Nucleic Acids Res.38: 1738-1748.
Galichet, A., K. Hoyerova, M. Kaminek, and W. Gruissem. 2008. Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol. 146: 1155-1164.
Gan, S. and R.M. Amasino. 1995. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986-1988.
Higuchi, M., M.S. Pischke, A.P. Mahonen, K. Miyawaki, Y. Hashimoto, M. Seki, M. Kobayashi, K. Shinozaki, T. Kato, S. Tabata, Y. Helariutta, M.R. Sussman, and T. Kakimoto. 2004. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101: 8821-8826.
Kakimoto, T. 2001. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopente-nyltransferases. Plant Cell Physiol. 42: 677-685.
Kasahara, H., K. Takei, N. Ueda, S. Hishiyama, T. Yamaya, Y. Kamiya, S. Yamaguchi, and H. Sakakibara. 2004. Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 279: 14049-14054.
Kim, H.J., H. Ryu, S.H. Hong, H.R. Woo, P.O. Lim, I.C. Lee,
J. Sheen, H.G. Nam, and I. Hwang. 2006. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation
|
of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 103:
814-819.
Mok, D.W. and M.C. Mok. 2001. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 89-118.
Murai, N., D.W.S. Mok, and M.C. Mok (eds.). 1994. Cytokinin
biosynthesis in tRNA and cytokinin incorporation
into plant RNA. CRC Press, Florida, 87 pp.
Sakakibara, H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431-449.
Sakakibara, H., H. Kasahara, N. Ueda, M. Kojima, K. Takei, S. Hishiyama, T. Asami, K. Okada, Y. Kamiya, T. Yamaya, and S. Yamaguchi. 2005. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosyn-thetic pathway in the host plant. Proc. Natl. Acad. Sci. USA 102: 9972-9977.
Sakamoto, T., H. Sakakibara, M. Kojima, Y. Yamamoto, H. Nagasaki, Y. Inukai, Y. Sato, and M. Matsuoka. 2006. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54-62.
Sakano, Y., Y. Okada, A. Matsunaga, T. Suwama, T. Kaneko, K. Ito, H. Noguchi, and I. Abe. 2004. Molecular cloning, expression, and characterization of adenylate isopentenyl-transferase from hop (Humulus lupulus L.). Phytochemistry 65: 2439-2446.
Takei, K., H. Sakakibara, and T. Sugiyama. 2001. Identification of genes encoding adenylate isopentenyltransferase, a cyto-kinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276: 26405-26410.
Vreman, H.J. and F. Skoog. 1972. Cytokinins in Pisum
transfer ribonucleic acid. Plant Physiol. 49: 848-49851.
|
|