Bot. Bull. Acad. Sin. (2003) 44: 285-289
Huang et al. — Fine mapping of the nuclear fertility restorer gene
Fine mapping of the nuclear fertility restorer gene for HL cytoplasmic male sterility in rice
Jingyu Huang1, Jun Hu1, Xin Xu2, Shaoqing Li1, Ping Yi1, Daichang Yang3, Fugang Ren1, Xuequn Liu1,.2, and Yingguo Zhu1,*
1The Key Laboratory of Ministry of Education for Plant Developmental Biology and Institute of Genetics, College of Life Sciences, Wuhan University, Wuhan, 430072, P. R. China
2College of Life Sciences and Chemistry, South-Central University for Ethnic Communities, Wuhan, 430074, P. R. China
3Ventria Bioscience 4110 Freeway, Sacramento, CA 95834, USA
(Received December 2, 2002; Accepted May 16, 2003)
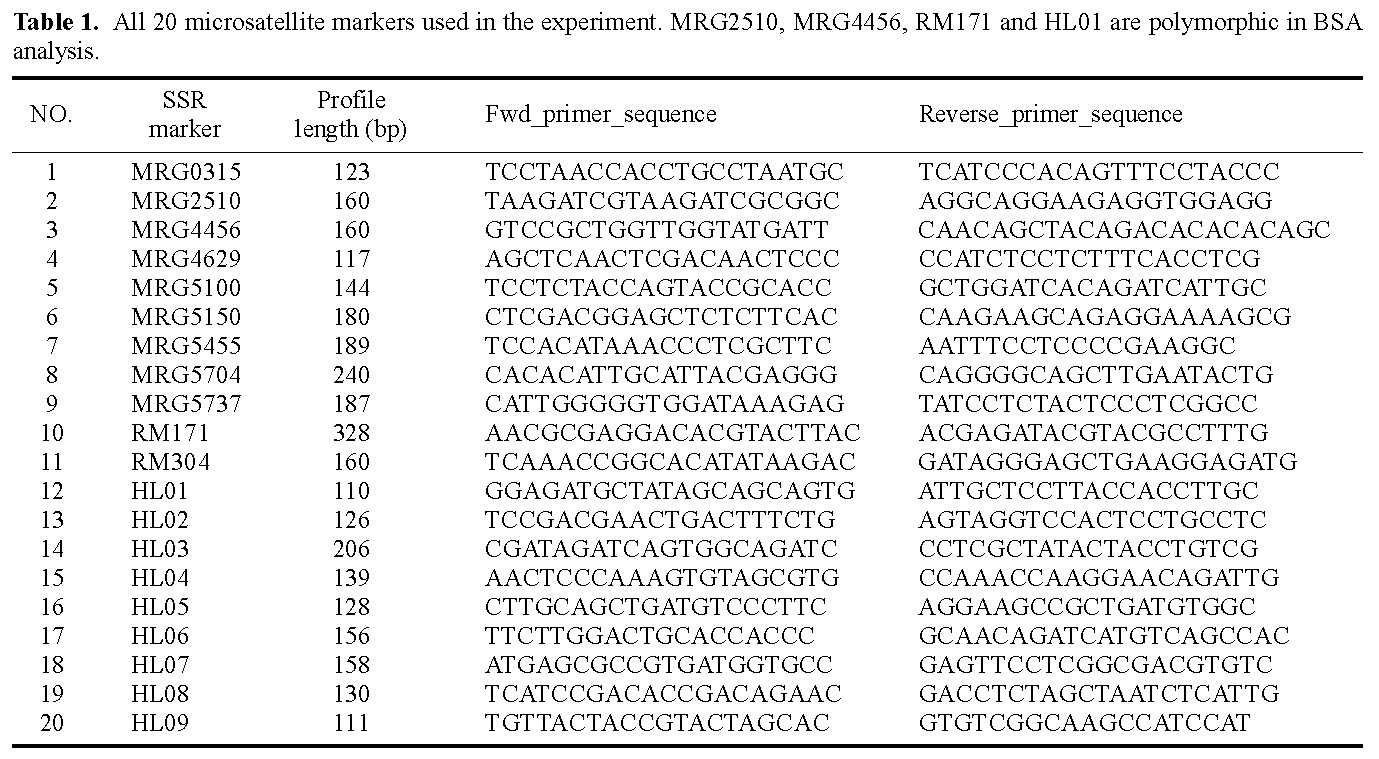
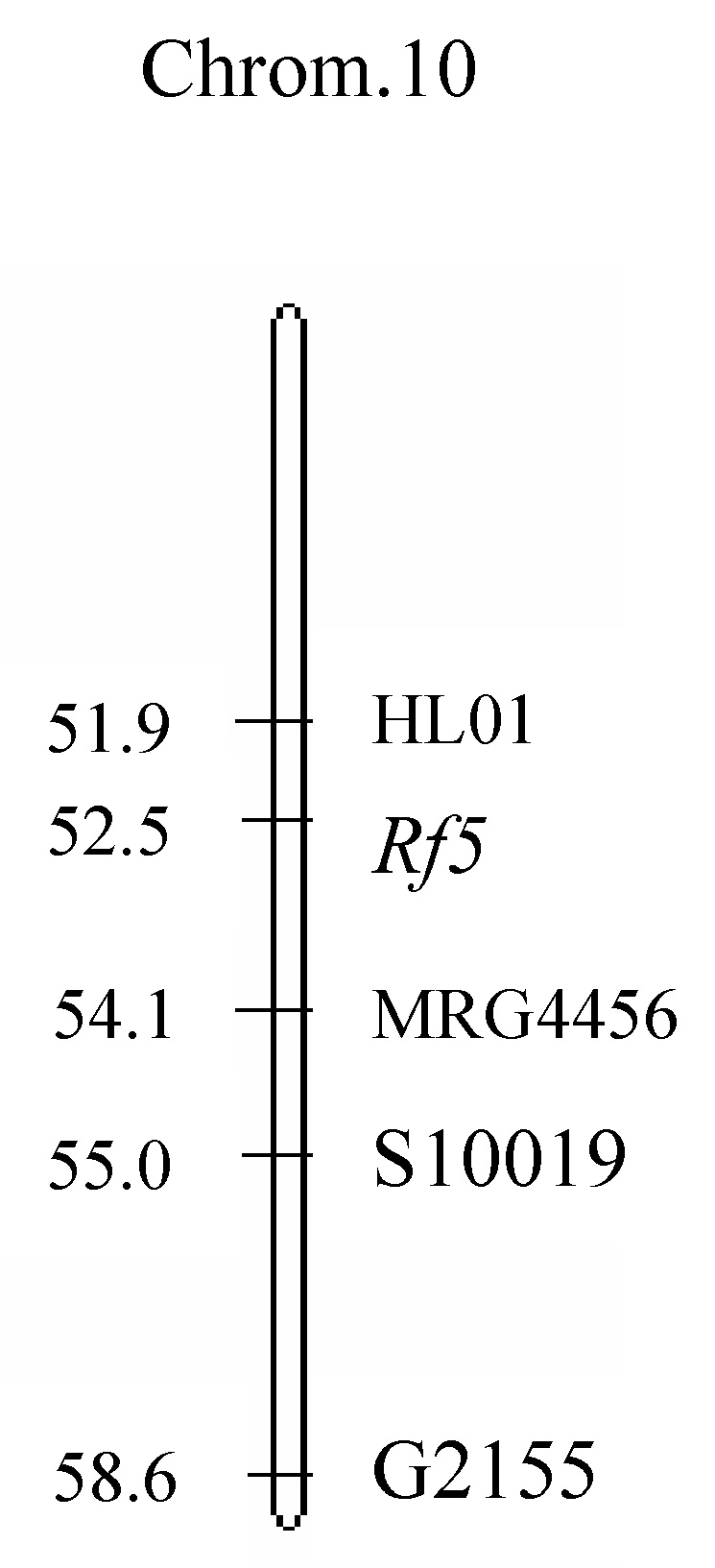
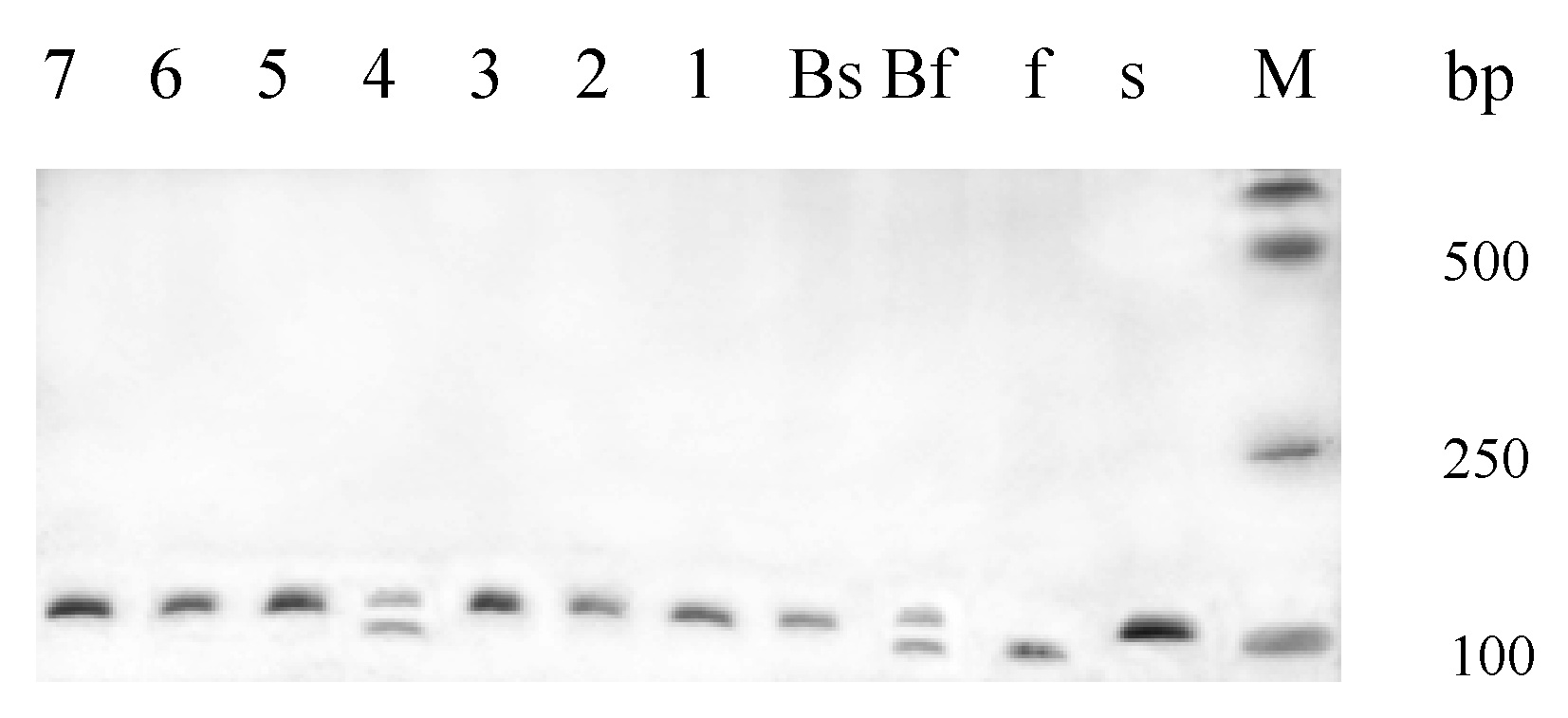
Abstract. Bulked segregant analysis (BSA) of a BC1 population derived from congguang 41A/MiYang 23//congguang 41B was used to map the nuclear fertility restorer gene Rf5 for HongLian (HL) cytoplasmic male sterility. The parents and two bulks representing extremely fertile and sterile plants, respectively, were screened for polymorphism with 20 microsatellite primer pairs on chromosome 10, chosen on the basis of previous research. MRG4456 is linked to the fertility restorer gene Rf5 at a distance of 1.57 cM, and another newly developed microsatellite primer, HL01, was locked at a distance of 0.63 cM to Rf5. Closely linked DNA markers will facilitate not only breeding but also the purity management of hybrid seeds. Concrete steps for developing new microsatellite markers using the rice whole genomic sequence database are also described.
Keywords: Fertility restorer gene; Marker-assisted selection; Microsatellite marker; Oryza sativa L.
Introduction
The phenomenon of cytoplasmic male sterility (CMS), described as the inability of a plant to produce functional pollens, has been observed in over 150 plant species (Wise et al., 2002). Fertility restorer (Rf) genes in the nuclear genome can counteract this inability (Newton, 1988) and restore fertility to cytoplasmic male-sterile plants. Hybrid breedings based on CMS/Rf systems have achieved great success all over the world. Aside from its commercial exploitation, CMS offers a rare opportunity to examine the regulation of mitochondrial genes by nuclear genes in multicellular organisms.
Three primary types of CMS in rice are now known, and their heritance habits and physiological characteristics have been extensively investigated. They are Wild-rice abortive (WA), BaoTai (BT) and HongLian (HL). WA type CMS belongs to sporophytic abortion, which fails to produce normal pollen and finally forms typical abortive pollen. In contrast, BT (japonica.) and HL type CMS (indica.) belong to gametic abortion, but they are also greatly different in terms of abortive phenotype, relationship of restoration, and maintenance.
CMS systems are usually attributed to chimeric ORFs in the mitochondrial genome (Kempken and Ping, 1998; Schnable et al., 1998; Szklarczyk et al., 2000). These ORFs encode novel proteins, which often interfere with mitochondrial function and pollen development. In many instances, the restorer gene suppression of CMS is directly associated with Rf-gene-dependent mitochondrial RNA modification and concurrent reduction of CMS associated protein (Schnabel et al., 1998). Although many mitochondrial genes associated with CMS have been cloned, only two Rf genes have been isolated until now. One is the maize Rf gene named Rf2, encoding aldehyde dehydrogenase (ALDH) (Cui et al., 1996; Liu et al., 2001); The other is Petunia Rf gene, encoding a mitochondrially targeted protein comprised of pentatricopeptide repeat (PPR) motif (Bentolila et al., 2002).
The inheritance of fertility restoration in WA type CMS has been extensively investigated. Most investigators tended to agree that restoration of WA type CMS is controlled by two nuclear genes (Rf3, Rf4) and that their chromosomal loci have been resolved (Zhang et al., 1997; Yao et al., 1997; Tan and Trangoonrang, 1998; Zhang et al., 2002). BT type CMS is restored by nuclear fertility restorer gene Rf1, which was mapped on chromosome 10 (Fukuta et al., 1992; Akagi et al., 1996;Yokozeki et al., 1996). HL type fertility restoration gene Rf5 was also mapped on chromosome 10, 7.8 cM from RM258 (Huang et al., 2000). The studies reported here were undertaken to finely locate the nuclear fertility restoration gene Rf5 for HL type CMS.
*Corresponding author. Tel: 86-27-87876530; Fax: 86-27-87876530; E-mail: Zhuyg@public.wh.hb.cn