Bot. Bull. Acad. Sin. (2004) 45: 33-40
Chen et al. — Evolution of apocarpy in Alismatidae
Evolution of apocarpy in Alismatidae using phylogenetic evidence from chloroplast rbcL gene sequence data
Jin-Ming Chen1, Dan Chen1, Wahiti Robert Gituru1.2, Qing-Feng Wang1,*, and You-Hao Guo1
1Laboratory of Plant Systematics and Evolutionary Biology, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072, P.R. China
2Present address: Botany Department, Kenyatta University, P. O. Box 43844, Nairobi, Kenya
(Received November 26, 2002; Accepted July 22, 2003)
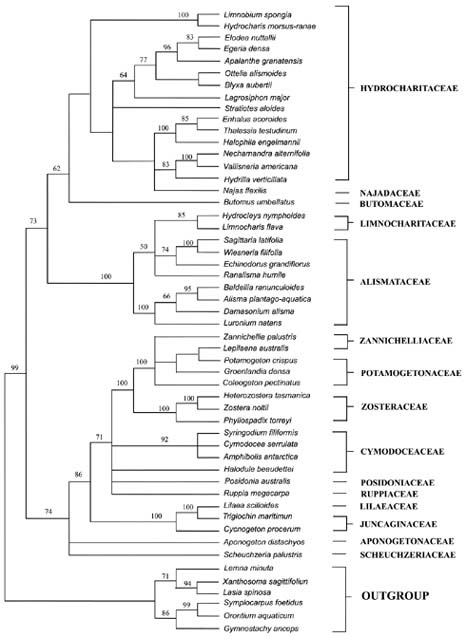
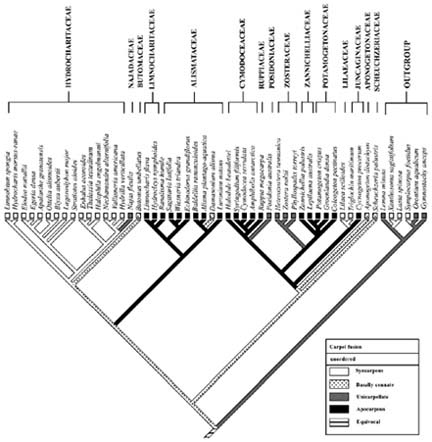
Abstract. The apocarpous groups in the monocotyledons are mainly concentrated in the subclass Alismatidae. The molecular phylogeny of Alismatidae based on analysis of chloroplast rbcL gene sequence data serves as a framework with which to evaluate character evolution with respect to the derivation of apocarpy in the group. 20 of the 27 genera in the subclass that display apocarpy have been included in our study. Our analysis indicates that apocarpy is polyphyletic within the subclass Alismatidae. Two independent origins of apocarpy in Alismatidae are explored in this study. Three separate origins of a single carpel and two separate origins of syncarpy in the subclass are also proposed. Basally connate carpel condition was the ancestral character in Alismatidae and evolved in two directions. It is possible for the unicarpellate condition to have been directly derived by reduction from syncarpy, and it could also be that the unicarpellate state has been derived from apocarpy by reduction in carpel number. The present results indicate that a progression has occurred in the evolution of carpels in Alismatidae from basally connate carpels through syncarpy or apocarpy to a single carpel.
Keywords: Alismatidae; Apocarpy; Basally connate carpels; RbcL gene; Single carpel; Syncarpy; Unicarpellate.
Introduction
Apocarpy has been regarded as an ancestral character in the angiosperms (Bessey, 1915; Hutchinson, 1959). Doyle and Endress (2000) concluded that apocarpy is a primitive feature in angiosperms, but that in monocotyledons it represents a reversal from syncarpy. The apocarpous groups in the monocotyledons include almost all clades of the subclass Alismatidae (Cronquist, 1981) or Alismatiflorae (Stebbins, 1974; Thorne, 1976; Dahlgren and Clifford, 1982). Previous researchers hypothesized that the apocarpous groups in the Alismatidae were the most primitive monocotyledons (Hutchinson, 1959; Takhtajan, 1980; Cronquist, 1988). The origin of the monocotyledons from the Ranunculaceae was strongly advocated by Hutchinson (1973), who relied on the feature of apocarpy as evidence of their relationship. Although apocarpy is generally believed to represent the ancestral condition in Alismatidae (Cronquist, 1981), it has also been interpreted as derived by this subclass (Dahlgren and Rasmussen, 1983; Dahlgren et al., 1985).
The evolution of apocarpy is one of the most interesting evolutionary events in the history of monocotyledons. A better understanding of this evolution may provide valuable insight into the adaptive evolution in flowering plants and also assist in resolving the relationships among the apocarpous groups within the angiosperms. Several inves
tigations into the origin and evolution of carpel form and structure have been reported (Taylor and Hickey, 1996; Igersheim et al., 2001). Owing to the sampling limitations in most studies, the evolutionary relationships of apocarpy and other gynoecium conditions, such as syncarpous, basally connate, and unicarpellate conditions in monocotyledons, are still obscure. Here, we present a phylogenetic analysis of chloroplast rbcL gene sequences for 46 species from 46 genera representing all currently recognized families and 81% of the genera in the subclass Alismatidae. Delimitation of families and genera are according to Cook (1990), Cronquist (1981), and Tomlinson (1982). Les et al. (1997) constructed a phylogeny of the subclass Alismatidae, and the origins and evolution of marine angiosperms (seagrasses) and hydrophily in the subclass were also estimated; however, for our purpose only one species from each genus was selected because the condition of carpel fusion was constant in each genus. In the present analysis, 20 of the 27 genera that display apocarpy in the subclass are included. The molecular phylogeny of Alismatidae is used as a framework on which to evaluate character evolution with respect to the evolution of apocarpy in the group.
Materials and Methods
Taxa Sampling
RbcL sequences for 46 species of Alismatidae that were reported previously in Genbank were obtained. One rbcL sequence of Lemna (Lemnaceae) and five sequences of
*Corresponding author. Tel: +86-27-87682869; Fax: +86-27-87682869; E-mail: wangqf97@hotmail.com